A New Natural Product Analog of Blasticidin S Reveals Cellular Uptake Facilitated by the NorA Multidrug Transporter
- PMID: 28373194
- PMCID: PMC5444127
- DOI: 10.1128/AAC.02635-16
A New Natural Product Analog of Blasticidin S Reveals Cellular Uptake Facilitated by the NorA Multidrug Transporter
Abstract
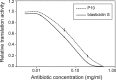
The permeation of antibiotics through bacterial membranes to their target site is a crucial determinant of drug activity but in many cases remains poorly understood. During screening efforts to discover new broad-spectrum antibiotic compounds from marine sponge samples, we identified a new analog of the peptidyl nucleoside antibiotic blasticidin S that exhibited up to 16-fold-improved potency against a range of laboratory and clinical bacterial strains which we named P10. Whole-genome sequencing of laboratory-evolved strains of Staphylococcus aureus resistant to blasticidin S and P10, combined with genome-wide assessment of the fitness of barcoded Escherichia coli knockout strains in the presence of the antibiotics, revealed that restriction of cellular access was a key feature in the development of resistance to this class of drug. In particular, the gene encoding the well-characterized multidrug efflux pump NorA was found to be mutated in 69% of all S. aureus isolates resistant to blasticidin S or P10. Unexpectedly, resistance was associated with inactivation of norA, suggesting that the NorA transporter facilitates cellular entry of peptidyl nucleosides in addition to its known role in the efflux of diverse compounds, including fluoroquinolone antibiotics.
Keywords: antibiotic resistance; chemical genomics; natural products; transporters.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, Lerner SA, Levy S, Lewis K, Lomovskaya O, Miller JH, Mobashery S, Piddock LJV, Projan S, Thomas CM, Tomasz A, Tulkens PM, Walsh TM, Watson JD, Witkowski J, Witte W, Wright G, Yeh P, Zgurskaya HI. 2011. Tackling antibiotic resistance. Nat Rev Microbiol 9:894–896. doi:10.1038/nrmicro2693. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

