Heat shock protein 104 (Hsp104)-mediated curing of [ PSI+] yeast prions depends on both [ PSI+] conformation and the properties of the Hsp104 homologs
- PMID: 28373280
- PMCID: PMC5448092
- DOI: 10.1074/jbc.M116.770719
Heat shock protein 104 (Hsp104)-mediated curing of [ PSI+] yeast prions depends on both [ PSI+] conformation and the properties of the Hsp104 homologs
Abstract
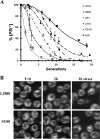
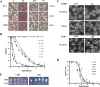
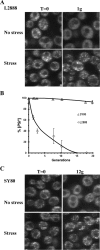
Prions arise from proteins that have two possible conformations: properly folded and non-infectious or misfolded and infectious. The [PSI+] yeast prion, which is the misfolded and self-propagating form of the translation termination factor eRF3 (Sup35), can be cured of its infectious conformation by overexpression of Hsp104, which helps dissolve the prion seeds. This dissolution depends on the trimming activity of Hsp104, which reduces the size of the prion seeds without increasing their number. To further understand the relationship between trimming and curing, trimming was followed by measuring the loss of GFP-labeled Sup35 foci from both strong and weak [PSI+] variants; the former variant has more seeds and less soluble Sup35 than the latter. Overexpression of Saccharomyces cerevisiae Hsp104 (Sc-Hsp104) trimmed the weak [PSI+] variants much faster than the strong variants and cured the weak variants an order of magnitude faster than the strong variants. Overexpression of the fungal Hsp104 homologs from Schizosaccharomyces pombe (Sp-Hsp104) or Candida albicans (Ca-Hsp104) also trimmed and cured the weak variants, but interestingly, it neither trimmed nor cured the strong variants. These results show that, because Sc-Hsp104 has greater trimming activity than either Ca-Hsp104 or Sp-Hsp104, it cures both the weak and strong variants, whereas Ca-Hsp104 and Sp-Hsp104 only cure the weak variants. Therefore, curing by Hsp104 overexpression depends on both the trimming ability of the fungal Hsp104 homolog and the strength of the [PSI+] variant: the greater the trimming activity of the Hsp104 homolog and the weaker the variant, the greater the curing.
Keywords: Hsp104; amyloid; molecular chaperone; prion; protein folding; yeast.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
Curing of [PSI+] by Hsp104 Overexpression: Clues to solving the puzzle.Prion. 2018 Jan 2;12(1):9-15. doi: 10.1080/19336896.2017.1412911. Epub 2018 Feb 2. Prion. 2018. PMID: 29227184 Free PMC article.
-
Hsp104 overexpression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds.Eukaryot Cell. 2014 May;13(5):635-47. doi: 10.1128/EC.00300-13. Epub 2014 Mar 14. Eukaryot Cell. 2014. PMID: 24632242 Free PMC article.
-
Overexpression of Hsp104 by Causing Dissolution of the Prion Seeds Cures the Yeast [PSI+] Prion.Int J Mol Sci. 2023 Jun 29;24(13):10833. doi: 10.3390/ijms241310833. Int J Mol Sci. 2023. PMID: 37446010 Free PMC article.
-
The life of [PSI].Curr Genet. 2018 Feb;64(1):1-8. doi: 10.1007/s00294-017-0714-7. Epub 2017 Jun 26. Curr Genet. 2018. PMID: 28653109 Free PMC article. Review.
-
Mechanisms for Curing Yeast Prions.Int J Mol Sci. 2020 Sep 7;21(18):6536. doi: 10.3390/ijms21186536. Int J Mol Sci. 2020. PMID: 32906758 Free PMC article. Review.
Cited by
-
Three J-proteins impact Hsp104-mediated variant-specific prion elimination: a new critical role for a low-complexity domain.Curr Genet. 2020 Feb;66(1):51-58. doi: 10.1007/s00294-019-01006-5. Epub 2019 Jun 22. Curr Genet. 2020. PMID: 31230108 Free PMC article. Review.
-
Thermotolerance improvement of engineered Saccharomyces cerevisiae ERG5 Delta ERG4 Delta ERG3 Delta, molecular mechanism, and its application in corn ethanol production.Biotechnol Biofuels Bioprod. 2023 Apr 12;16(1):66. doi: 10.1186/s13068-023-02312-4. Biotechnol Biofuels Bioprod. 2023. PMID: 37046321 Free PMC article.
-
Total propagation of yeast prion conformers in ssz1∆ upf1∆ Hsp104T160M triple mutants.Curr Genet. 2025 Mar 29;71(1):8. doi: 10.1007/s00294-025-01313-0. Curr Genet. 2025. PMID: 40156734 Free PMC article.
-
The Cryo-EM Effect: Structural Biology of Neurodegenerative Disease Proteostasis Factors.J Neuropathol Exp Neurol. 2021 Jun 4;80(6):494-513. doi: 10.1093/jnen/nlab029. J Neuropathol Exp Neurol. 2021. PMID: 33860329 Free PMC article. Review.
-
Transcriptional profiling provides new insights into the role of nitric oxide in enhancing Ganoderma oregonense resistance to heat stress.Sci Rep. 2017 Nov 16;7(1):15694. doi: 10.1038/s41598-017-15340-6. Sci Rep. 2017. PMID: 29146915 Free PMC article.
References
-
- Doyle S. M., and Wickner S. (2009) Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 34, 40–48 - PubMed
-
- Ferreira P. C., Ness F., Edwards S. R., Cox B. S., and Tuite M. F. (2001) The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40, 1357–1369 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

