T Cells in Celiac Disease
- PMID: 28373482
- PMCID: PMC5426360
- DOI: 10.4049/jimmunol.1601693
T Cells in Celiac Disease
Abstract
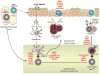
Celiac disease is a human T cell-mediated autoimmune-like disorder caused by exposure to dietary gluten in genetically predisposed individuals. This review will discuss how CD4 T cell responses directed against an exogenous Ag can cause an autoreactive B cell response and participate in the licensing of intraepithelial lymphocytes to kill intestinal epithelial cells. Furthermore, this review will examine the mechanisms by which intraepithelial cytotoxic T cells mediate tissue destruction in celiac disease.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures


References
-
- Sollid LM. Molecular basis of celiac disease. Annu Rev Immunol. 2000;18:53–81. - PubMed
-
- Abadie V, Sollid LM, Barreiro LB, Jabri B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol. 2011;29:493–525. - PubMed
-
- Hunt KA, van Heel DA. Recent advances in coeliac disease genetics. Gut. 2009;58:473–476. - PubMed
-
- Withoff S, Li Y, Jonkers I, Wijmenga C. Understanding Celiac Disease by Genomics. Trends in genetics : TIG. 2016;32:295–308. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

