Emergence of a new Neisseria meningitidis clonal complex 11 lineage 11.2 clade as an effective urogenital pathogen
- PMID: 28373547
- PMCID: PMC5402416
- DOI: 10.1073/pnas.1620971114
Emergence of a new Neisseria meningitidis clonal complex 11 lineage 11.2 clade as an effective urogenital pathogen
Abstract
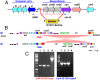
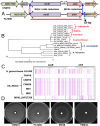
Neisseria meningitidis (Nm) clonal complex 11 (cc11) lineage is a hypervirulent pathogen responsible for outbreaks of invasive meningococcal disease, including among men who have sex with men, and is increasingly associated with urogenital infections. Recently, clusters of Nm urethritis have emerged primarily among heterosexual males in the United States. We determined that nonencapsulated meningococcal isolates from an ongoing Nm urethritis outbreak among epidemiologically unrelated men in Columbus, Ohio, are linked to increased Nm urethritis cases in multiple US cities, including Atlanta and Indianapolis, and that they form a unique clade (the US Nm urethritis clade, US_NmUC). The isolates belonged to the cc11 lineage 11.2/ET-15 with fine type of PorA P1.5-1, 10-8; FetA F3-6; PorB 2-2 and express a unique FHbp allele. A common molecular fingerprint of US_NmUC isolates was an IS1301 element in the intergenic region separating the capsule ctr-css operons and adjacent deletion of cssA/B/C and a part of csc, encoding the serogroup C capsule polymerase. This resulted in the loss of encapsulation and intrinsic lipooligosaccharide sialylation that may promote adherence to mucosal surfaces. Furthermore, we detected an IS1301-mediated inversion of an ∼20-kb sequence near the cps locus. Surprisingly, these isolates had acquired by gene conversion the complete gonococcal denitrification norB-aniA gene cassette, and strains grow well anaerobically. The cc11 US_NmUC isolates causing urethritis clusters in the United States may have adapted to a urogenital environment by loss of capsule and gene conversion of the Neisseria gonorrheae norB-aniA cassette promoting anaerobic growth.
Keywords: IS1301; Neisseria meningitidis; capsule; denitrification; meningococcal urethritis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
Emergence of a Urogenital Pathotype of Neisseria meningitidis.Trends Microbiol. 2017 Jul;25(7):510-512. doi: 10.1016/j.tim.2017.05.006. Epub 2017 May 22. Trends Microbiol. 2017. PMID: 28545723
References
-
- Wilder-Smith A. Meningococcal vaccine in travelers. Curr Opin Infect Dis. 2007;20:454–460. - PubMed
-
- William DC, Schapiro CM, Felman YM. Pharyngeal carriage of Neisseria meningitidis and anogenital gonorrhea: Evidence for their relationship. Sex Transm Dis. 1980;7:175–177. - PubMed
-
- Janda WM, Bohnoff M, Morello JA, Lerner SA. Prevalence and site-pathogen studies of Neisseria meningitidis and N gonorrhoeae in homosexual men. JAMA. 1980;244:2060–2064. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

