CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome
- PMID: 28373564
- PMCID: PMC5402455
- DOI: 10.1073/pnas.1700530114
CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome
Abstract
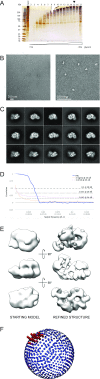
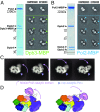
The replisome unwinds and synthesizes DNA for genome duplication. In eukaryotes, the Cdc45-MCM-GINS (CMG) helicase and the leading-strand polymerase, Pol epsilon, form a stable assembly. The mechanism for coupling DNA unwinding with synthesis is starting to be elucidated, however the architecture and dynamics of the replication fork remain only partially understood, preventing a molecular understanding of chromosome replication. To address this issue, we conducted a systematic single-particle EM study on multiple permutations of the reconstituted CMG-Pol epsilon assembly. Pol epsilon contains two flexibly tethered lobes. The noncatalytic lobe is anchored to the motor of the helicase, whereas the polymerization domain extends toward the side of the helicase. We observe two alternate configurations of the DNA synthesis domain in the CMG-bound Pol epsilon. We propose that this conformational switch might control DNA template engagement and release, modulating replisome progression.
Keywords: CMG helicase; DNA polymerase; DNA replication; single-particle electron microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures













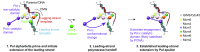
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

