Single-Molecule Studies of Telomeres and Telomerase
- PMID: 28375735
- PMCID: PMC5624789
- DOI: 10.1146/annurev-biophys-062215-011256
Single-Molecule Studies of Telomeres and Telomerase
Abstract
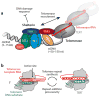
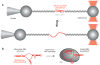
Telomeres are specialized chromatin structures that protect chromosome ends from dangerous processing events. In most tissues, telomeres shorten with each round of cell division, placing a finite limit on cell growth. In rapidly dividing cells, including the majority of human cancers, cells bypass this growth limit through telomerase-catalyzed maintenance of telomere length. The dynamic properties of telomeres and telomerase render them difficult to study using ensemble biochemical and structural techniques. This review describes single-molecule approaches to studying how individual components of telomeres and telomerase contribute to function. Single-molecule methods provide a window into the complex nature of telomeres and telomerase by permitting researchers to directly visualize and manipulate the individual protein, DNA, and RNA molecules required for telomere function. The work reviewed in this article highlights how single-molecule techniques have been utilized to investigate the function of telomeres and telomerase.
Keywords: single-molecule biophysics; telomerase; telomere.
Figures





References
-
- Alves D, Li H, Codrington R, Orte A, Ren X, et al. Single-molecule analysis of human telomerase monomer. Nat Chem Biol. 2008;4:287–89. - PubMed
-
- Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol. 2007;14:147–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

