Application Value of Mass Spectrometry in the Differentiation of Benign and Malignant Liver Tumors
- PMID: 28376075
- PMCID: PMC5388305
- DOI: 10.12659/msm.901064
Application Value of Mass Spectrometry in the Differentiation of Benign and Malignant Liver Tumors
Abstract
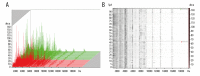
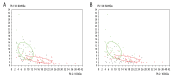
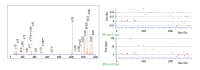
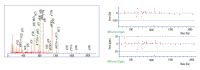
BACKGROUND Differentiation of malignant from benign liver tumors remains a challenging problem. In recent years, mass spectrometry (MS) technique has emerged as a promising strategy to diagnose a wide range of malignant tumors. The purpose of this study was to establish classification models to distinguish benign and malignant liver tumors and identify the liver cancer-specific peptides by mass spectrometry. MATERIAL AND METHODS In our study, serum samples from 43 patients with malignant liver tumors and 52 patients with benign liver tumors were treated with weak cation-exchange chromatography Magnetic Beads (MB-WCX) kits and analyzed by the Matrix-Assisted Laser Desorption Time of Flight Mass Spectrometry (MALDI-TOF-MS). Then we established genetic algorithm (GA), supervised neural networks (SNN), and quick classifier (QC) models to distinguish malignant from benign liver tumors. To confirm the clinical applicability of the established models, the blinded validation test was performed in 50 clinical serum samples. Discriminatory peaks associated with malignant liver tumors were subsequently identified by a qTOF Synapt G2-S system. RESULTS A total of 27 discriminant peaks (p<0.05) in mass spectra of serum samples were found by ClinPro Tools software. Recognition capabilities of the established models were 100% (GA), 89.38% (SNN), and 80.84% (QC); cross-validation rates were 81.67% (GA), 81.11% (SNN), and 86.11% (QC). The accuracy rates of the blinded validation test were 78% (GA), 84% (SNN), and 84% (QC). From the 27 discriminatory peptide peaks analyzed, 3 peaks of m/z 2860.34, 2881.54, and 3155.67 were identified as a fragment of fibrinogen alpha chain, fibrinogen beta chain, and inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4), respectively. CONCLUSIONS Our results demonstrated that MS technique can be helpful in differentiation of benign and malignant liver tumors. Fibrinogen and ITIH4 might be used as biomarkers for the diagnosis of malignant liver tumors.
Figures

Similar articles
-
Detection and Identification of Serum Peptides Biomarker in Papillary Thyroid Cancer.Med Sci Monit. 2018 Mar 17;24:1581-1587. doi: 10.12659/msm.907768. Med Sci Monit. 2018. PMID: 29549708 Free PMC article.
-
Matrix-assisted laser desorption ionization: time of flight mass spectrometry-identified models for detection of ESBL-producing bacterial strains.Med Sci Monit Basic Res. 2014 Nov 12;20:176-83. doi: 10.12659/MSMBR.892670. Med Sci Monit Basic Res. 2014. PMID: 25390932 Free PMC article.
-
Mass spectrometry-based serum peptide profiling in hepatocellular carcinoma with bone metastasis.World J Gastroenterol. 2014 Mar 21;20(11):3025-32. doi: 10.3748/wjg.v20.i11.3025. World J Gastroenterol. 2014. PMID: 24659894 Free PMC article.
-
Serum peptidome patterns of hepatocellular carcinoma based on magnetic bead separation and mass spectrometry analysis.Diagn Pathol. 2013 Aug 5;8:130. doi: 10.1186/1746-1596-8-130. Diagn Pathol. 2013. PMID: 23915185 Free PMC article.
-
Application of serum protein fingerprinting coupled with artificial neural network model in diagnosis of hepatocellular carcinoma.Chin Med J (Engl). 2005 Aug 5;118(15):1278-84. Chin Med J (Engl). 2005. PMID: 16117882 Clinical Trial.
Cited by
-
A Fibrinogen Alpha Fragment Mitigates Chemotherapy-Induced MLL Rearrangements.Front Oncol. 2021 Jun 18;11:689063. doi: 10.3389/fonc.2021.689063. eCollection 2021. Front Oncol. 2021. PMID: 34222016 Free PMC article.
-
Identification of MST1 as a potential early detection biomarker for colorectal cancer through a proteomic approach.Sci Rep. 2017 Oct 27;7(1):14265. doi: 10.1038/s41598-017-14539-x. Sci Rep. 2017. PMID: 29079854 Free PMC article.
-
Detection and Identification of Serum Peptides Biomarker in Papillary Thyroid Cancer.Med Sci Monit. 2018 Mar 17;24:1581-1587. doi: 10.12659/msm.907768. Med Sci Monit. 2018. PMID: 29549708 Free PMC article.
-
Serum proteomic predicts effectiveness and reveals potential biomarkers for complications in liver transplant patients.Aging (Albany NY). 2020 Jun 12;12(12):12119-12141. doi: 10.18632/aging.103381. Epub 2020 Jun 12. Aging (Albany NY). 2020. PMID: 32530819 Free PMC article.
-
MALDI-TOF-MS analysis in low molecular weight serum peptidome biomarkers for NSCLC.J Clin Lab Anal. 2022 Apr;36(4):e24254. doi: 10.1002/jcla.24254. Epub 2022 Feb 25. J Clin Lab Anal. 2022. PMID: 35212031 Free PMC article.
References
-
- El Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–76. - PubMed
-
- Tian L, Wang Y, Xu D, et al. Serological AFP/Golgi protein 73 could be a new diagnostic parameter of hepatic diseases. Int J Cancer. 2011;129(8):1923–31. - PubMed
-
- Özmen E, Adaletli I, Kayadibi Y, et al. The impact of share wave elastography in differentiation of hepatic hemangioma from malignant liver tumors in pediatric population. Eur J Radiol. 2014;83(9):1691–97. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

