Co-activation of STAT3 and YES-Associated Protein 1 (YAP1) Pathway in EGFR-Mutant NSCLC
- PMID: 28376152
- PMCID: PMC5409000
- DOI: 10.1093/jnci/djx014
Co-activation of STAT3 and YES-Associated Protein 1 (YAP1) Pathway in EGFR-Mutant NSCLC
Abstract
Background: The efficacy of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in EGFR-mutant non-small cell lung cancer (NSCLC) is limited by adaptive activation of cell survival signals. We hypothesized that both signal transducer and activator of transcription 3 (STAT3) and Src-YES-associated protein 1 (YAP1) signaling are dually activated during EGFR TKI treatment to limit therapeutic response.
Methods: We used MTT and clonogenic assays, immunoblotting, and quantitative polymerase chain reaction to evaluate the efficacy of EGFR TKI alone and in combination with STAT3 and Src inhibition in three EGFR-mutant NSCLC cell lines. The Chou-Talalay method was used for the quantitative determination of drug interaction. We examined tumor growth inhibition in one EGFR-mutant NSCLC xenograft model (n = 4 mice per group). STAT3 and YAP1 expression was evaluated in tumors from 119 EGFR-mutant NSCLC patients (64 in an initial cohort and 55 in a validation cohort) by quantitative polymerase chain reaction. Kaplan-Meier and Cox regression analyses were used to assess the correlation between survival and gene expression. All statistical tests were two-sided.
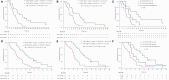
Results: We discovered that lung cancer cells survive initial EGFR inhibitor treatment through activation of not only STAT3 but also Src-YAP1 signaling. Cotargeting EGFR, STAT3, and Src was synergistic in two EGFR-mutant NSCLC cell lines with a combination index of 0.59 (95% confidence interval [CI] = 0.54 to 0.63) for the PC-9 and 0.59 (95% CI = 0.54 to 0.63) for the H1975 cell line. High expression of STAT3 or YAP1 predicted worse progression-free survival (hazard ratio [HR] = 3.02, 95% CI = 1.54 to 5.93, P = .001, and HR = 2.57, 95% CI = 1.30 to 5.09, P = .007, respectively) in an initial cohort of 64 EGFR-mutant NSCLC patients treated with firstline EGFR TKIs. Similar results were observed in a validation cohort.
Conclusions: Our study uncovers a coordinated signaling network centered on both STAT3 and Src-YAP signaling that limits targeted therapy response in lung cancer and identifies an unforeseen rational upfront polytherapy strategy to minimize residual disease and enhance clinical outcomes.
© The Author 2017. Published by Oxford University Press.
Figures





References
-
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–967. - PubMed
-
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. - PubMed
-
- Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305(5687):1163–1167. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

