Tumor Sequencing and Patient-Derived Xenografts in the Neoadjuvant Treatment of Breast Cancer
- PMID: 28376176
- PMCID: PMC5408989
- DOI: 10.1093/jnci/djw306
Tumor Sequencing and Patient-Derived Xenografts in the Neoadjuvant Treatment of Breast Cancer
Abstract
Background: Breast cancer patients with residual disease after neoadjuvant chemotherapy (NAC) have increased recurrence risk. Molecular characterization, knowledge of NAC response, and simultaneous generation of patient-derived xenografts (PDXs) may accelerate drug development. However, the feasibility of this approach is unknown.
Methods: We conducted a prospective study of 140 breast cancer patients treated with NAC and performed tumor and germline sequencing and generated patient-derived xenografts (PDXs) using core needle biopsies. Chemotherapy response was assessed at surgery.
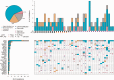
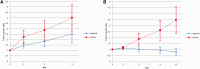
Results: Recurrent "targetable" alterations were not enriched in patients without pathologic complete response (pCR); however, upregulation of steroid receptor signaling and lower pCR rates (16.7%, 1/6) were observed in triple-negative breast cancer (TNBC) patients with luminal androgen receptor (LAR) vs basal subtypes (60.0%, 21/35). Within TNBC, TP53 mutation frequency (75.6%, 31/41) did not differ comparing basal (74.3%, 26/35) and LAR (83.3%, 5/6); however, TP53 stop-gain mutations were more common in basal (22.9%, 8/35) vs LAR (0.0%, 0/6), which was confirmed in The Cancer Genome Atlas and British Columbia data sets. In luminal B tumors, Ki-67 responses were observed in tumors that harbored mutations conferring endocrine resistance ( p53, AKT, and IKBKE ). PDX take rate (27.4%, 31/113) varied according to tumor subtype, and in a patient with progression on NAC, sequencing data informed drug selection (olaparib) with in vivo antitumor activity observed in the primary and resistant (postchemotherapy) PDXs.
Conclusions: In this study, we demonstrate the feasibility of tumor sequencing and PDX generation in the NAC setting. "Targetable" alterations were not enriched in chemotherapy-resistant tumors; however, prioritization of drug testing based on sequence data may accelerate drug development.
© The Author, 2017. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com
Figures

Comment in
-
Breast Cancer Patient-Derived Xenografts: Pros, Cons, and Next Steps.J Natl Cancer Inst. 2017 Jul 1;109(7):djw307. doi: 10.1093/jnci/djw307. J Natl Cancer Inst. 2017. PMID: 28376181 Free PMC article. No abstract available.
References
-
- Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

