Combination Therapy Targeting BCL6 and Phospho-STAT3 Defeats Intratumor Heterogeneity in a Subset of Non-Small Cell Lung Cancers
- PMID: 28377453
- PMCID: PMC5489259
- DOI: 10.1158/0008-5472.CAN-15-3052
Combination Therapy Targeting BCL6 and Phospho-STAT3 Defeats Intratumor Heterogeneity in a Subset of Non-Small Cell Lung Cancers
Abstract
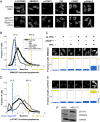
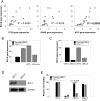
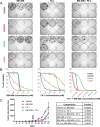
Oncogene-specific changes in cellular signaling have been widely observed in lung cancer. Here, we investigated how these alterations could affect signaling heterogeneity and suggest novel therapeutic strategies. We compared signaling changes across six human bronchial epithelial cell (HBEC) strains that were systematically transformed with various combinations of TP53, KRAS, and MYC-oncogenic alterations commonly found in non-small cell lung cancer (NSCLC). We interrogated at single-cell resolution how these alterations could affect classic readouts (β-CATENIN, SMAD2/3, phospho-STAT3, P65, FOXO1, and phospho-ERK1/2) of key pathways commonly affected in NSCLC. All three oncogenic alterations were required concurrently to observe significant signaling changes, and significant heterogeneity arose in this condition. Unexpectedly, we found two mutually exclusive altered subpopulations: one with STAT3 upregulation and another with SMAD2/3 downregulation. Treatment with a STAT3 inhibitor eliminated the upregulated STAT3 subpopulation, but left a large surviving subpopulation with downregulated SMAD2/3. A bioinformatics search identified BCL6, a gene downstream of SMAD2/3, as a novel pharmacologically accessible target of our transformed HBECs. Combination treatment with STAT3 and BCL6 inhibitors across a panel of NSCLC cell lines and in xenografted tumors significantly reduced tumor cell growth. We conclude that BCL6 is a new therapeutic target in NSCLC and combination therapy that targets multiple vulnerabilities (STAT3 and BCL6) downstream of common oncogenes, and tumor suppressors may provide a potent way to defeat intratumor heterogeneity. Cancer Res; 77(11); 3070-81. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

