GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects
- PMID: 28377535
- PMCID: PMC5656050
- DOI: 10.1136/jmedgenet-2016-104509
GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects
Abstract
Background: We aimed for a comprehensive delineation of genetic, functional and phenotypic aspects of GRIN2B encephalopathy and explored potential prospects of personalised medicine.
Methods: Data of 48 individuals with de novo GRIN2B variants were collected from several diagnostic and research cohorts, as well as from 43 patients from the literature. Functional consequences and response to memantine treatment were investigated in vitro and eventually translated into patient care.
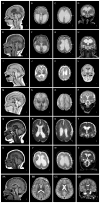
Results: Overall, de novo variants in 86 patients were classified as pathogenic/likely pathogenic. Patients presented with neurodevelopmental disorders and a spectrum of hypotonia, movement disorder, cortical visual impairment, cerebral volume loss and epilepsy. Six patients presented with a consistent malformation of cortical development (MCD) intermediate between tubulinopathies and polymicrogyria. Missense variants cluster in transmembrane segments and ligand-binding sites. Functional consequences of variants were diverse, revealing various potential gain-of-function and loss-of-function mechanisms and a retained sensitivity to the use-dependent blocker memantine. However, an objectifiable beneficial treatment response in the respective patients still remains to be demonstrated.
Conclusions: In addition to previously known features of intellectual disability, epilepsy and autism, we found evidence that GRIN2B encephalopathy is also frequently associated with movement disorder, cortical visual impairment and MCD revealing novel phenotypic consequences of channelopathies.
Keywords: channelopathy; clustering of missense variants; epileptic encephalopathy; pathogenic GRIN2B mutations; precision medicine.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2017. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: SFT is a consultant of Janssen Pharmaceuticals, Inc., Pfizer Inc., Boehringer Ingelheim Pharma GmbH & Co. KG, and co-founder of NeurOp Inc.
Figures
References
-
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400. - PubMed
-
- Burnashev N, Szepetowski P. NMDA receptor subunit mutations in neurodevelopmental disorders. Curr Opin Pharmacol. 2015;20:73–82. - PubMed
-
- Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, Milh M, Kortüm F, Fritsch A, Pientka FK, Hellenbroich Y, Kalscheuer VM, Kohlhase J, Moog U, Rappold G, Rauch A, Ropers HH, von Spiczak S, Tönnies H, Villeneuve N, Villard L, Zabel B, Zenker M, Laube B, Reis A, Wieczorek D, Van Maldergem L, Kutsche K. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–6. - PubMed
-
- Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, Park AR, Spiegelman D, Dobrzeniecka S, Piton A, Tomitori H, Daoud H, Massicotte C, Henrion E, Diallo O, Shekarabi M, Marineau C, Shevell M, Maranda B, Mitchell G, Nadeau A, D’Anjou G, Vanasse M, Srour M, Lafrenière RG, Drapeau P, Lacaille JC, Kim E, Lee JR, Igarashi K, Huganir RL, Rouleau GA, Michaud JL S2D Group. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–16. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
