Flow Cytometric Measurement of Blood Cells with BCR-ABL1 Fusion Protein in Chronic Myeloid Leukemia
- PMID: 28377570
- PMCID: PMC5429594
- DOI: 10.1038/s41598-017-00755-y
Flow Cytometric Measurement of Blood Cells with BCR-ABL1 Fusion Protein in Chronic Myeloid Leukemia
Abstract
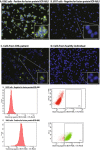
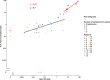
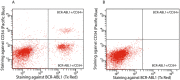
Chronic myeloid leukemia (CML) is characterized in the majority of cases by a t(9;22)(q34;q11) translocation, also called the Philadelphia chromosome, giving rise to the BCR-ABL1 fusion protein. Current treatment with tyrosine kinase inhibitors is directed against the constitutively active ABL1 domain of the fusion protein, and minimal residual disease (MRD) after therapy is monitored by real-time quantitative PCR (RQ-PCR) of the fusion transcript. Here, we describe a novel approach to detect and enumerate cells positive for the BCR-ABL1 fusion protein by combining the in situ proximity ligation assay with flow cytometry as readout (PLA-flow). By targeting of the BCR and ABL1 parts of the fusion protein with one antibody each, and creating strong fluorescent signals through rolling circle amplification, PLA-flow allowed sensitive detection of cells positive for the BCR-ABL1 fusion at frequencies as low as one in 10,000. Importantly, the flow cytometric results correlated strongly to those of RQ-PCR, both in diagnostic testing and for MRD measurements over time. In summary, we believe this flow cytometry-based method can serve as an attractive approach for routine measurement of cells harboring BCR-ABL1 fusions, also allowing simultaneously assessment of other cell surface markers as well as sensitive longitudinal follow-up.
Conflict of interest statement
UL is founder and holds stock in Olink Bioscience, having rights to the
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

