Rotiferan Hox genes give new insights into the evolution of metazoan bodyplans
- PMID: 28377584
- PMCID: PMC5431905
- DOI: 10.1038/s41467-017-00020-w
Rotiferan Hox genes give new insights into the evolution of metazoan bodyplans
Abstract
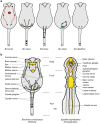
The phylum Rotifera consists of minuscule, nonsegmented animals with a unique body plan and an unresolved phylogenetic position. The presence of pharyngeal articulated jaws supports an inclusion in Gnathifera nested in the Spiralia. Comparison of Hox genes, involved in animal body plan patterning, can be used to infer phylogenetic relationships. Here, we report the expression of five Hox genes during embryogenesis of the rotifer Brachionus manjavacas and show how these genes define different functional components of the nervous system and not the usual bilaterian staggered expression along the anteroposterior axis. Sequence analysis revealed that the lox5-parapeptide, a key signature in lophotrochozoan and platyhelminthean Hox6/lox5 genes, is absent and replaced by different signatures in Rotifera and Chaetognatha, and that the MedPost gene, until now unique to Chaetognatha, is also present in rotifers. Collectively, our results support an inclusion of chaetognaths in gnathiferans and Gnathifera as sister group to the remaining spiralians.Rotifers are microscopic animals with an unusual, nonsegmented body plan consisting of a head, trunk and foot. Here, Fröbius and Funch investigate the role of Hox genes-which are widely used in animal body plan patterning-in rotifer embryogenesis and find non-canonical expression in the nervous system.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

