Microbial community compositions in the gastrointestinal tract of Chinese Mongolian sheep using Illumina MiSeq sequencing revealed high microbial diversity
- PMID: 28378284
- PMCID: PMC5380569
- DOI: 10.1186/s13568-017-0378-1
Microbial community compositions in the gastrointestinal tract of Chinese Mongolian sheep using Illumina MiSeq sequencing revealed high microbial diversity
Abstract
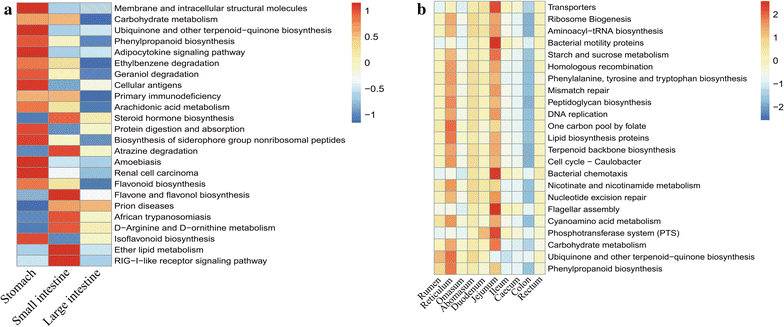
Chinese Mongolian sheep are an important ruminant raised for wool and meat production. However, little is known about the microbiota of the gastrointestinal tract (GIT) of Chinese Mongolian sheep. To increase our understanding of the microbial community composition in the GIT of Chinese Mongolian sheep, microbiota of five sheep is investigate for the first time using the Illumina MiSeq platform. High microbial diversity was obtained from the GIT, and the microbiota exhibited a higher biodiversity in the stomach and large intestine than in the small intestine. Firmicutes (44.62%), Bacteroidetes (38.49%), and Proteobacteria (4.11%) were the three most abundant phyla present in the GIT of the sheep. The present study also revealed the core genera of Prevotella, Bacteroides, Ruminococcus, Oscillospira, Treponema, and Desulfovibrio in the GIT. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States indicated that the metabolic pathway related to carbohydrate metabolism was the richest in the sheep GIT. In addition, a series of metabolic pathways related to plant secondary metabolism was most abundant in the stomach and large intestine than in the small intestine. Overall, the present study provides insight into the microbial community composition in GIT of the Chinese Mongolian sheep which is highly diverse and needs to be studied further to exploit the complex interactions with the host.
Keywords: Chinese Mongolian sheep; Gastrointestinal tract; Illumina MiSeq; Metabolic pathways; Microbiota.
Figures




References
-
- Arumugam M, Jeroen R, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin JJ, Sicheritzponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Consortium M. Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. - DOI - PMC - PubMed
-
- Avguštin G, Wallace RJ, Flint HJ. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int J Syst Bacteriol. 1997;2:284–288. doi: 10.1099/00207713-47-2-284. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

