A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer
- PMID: 28378740
- PMCID: PMC5382276
- DOI: 10.1038/ncomms14802
A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer
Abstract
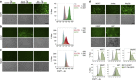
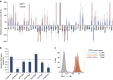
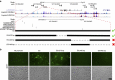
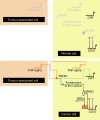
CD47 is a cell surface molecule that inhibits phagocytosis of cells that express it by binding to its receptor, SIRPα, on macrophages and other immune cells. CD47 is expressed at different levels by neoplastic and normal cells. Here, to reveal mechanisms by which different neoplastic cells generate this dominant 'don't eat me' signal, we analyse the CD47 regulatory genomic landscape. We identify two distinct super-enhancers (SEs) associated with CD47 in certain cancer cell types. We show that a set of active constituent enhancers, located within the two CD47 SEs, regulate CD47 expression in different cancer cell types and that disruption of CD47 SEs reduces CD47 gene expression. Finally we report that the TNF-NFKB1 signalling pathway directly regulates CD47 by interacting with a constituent enhancer located within a CD47-associated SE specific to breast cancer. These results suggest that cancers can evolve SE to drive CD47 overexpression to escape immune surveillance.
Conflict of interest statement
I.L.W. is co-founder, holds equity and is a Director and consultant of Forty Seven, Inc, a company developing CD47 antibody therapies. All other authors declare no competing financial interests.
Figures



Comment in
-
Inflammation, phagocytosis and cancer: another step in the CD47 act.J Thorac Dis. 2017 Aug;9(8):2279-2282. doi: 10.21037/jtd.2017.07.38. J Thorac Dis. 2017. PMID: 28932524 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

