Global evolutionary history and spatio-temporal dynamics of dengue virus type 2
- PMID: 28378782
- PMCID: PMC5381229
- DOI: 10.1038/srep45505
Global evolutionary history and spatio-temporal dynamics of dengue virus type 2
Abstract
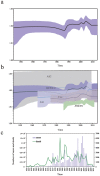
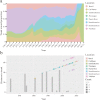
DENV-2 spread throughout the tropical and subtropical regions globally, which is implicated in deadly outbreaks of DHF and DSS. Since dengue cases have grown dramatically in recent years, about half of the world's population is now at risk. Our timescale analysis indicated that the most recent common ancestor existed about 100 years ago. The rate of nucleotide substitution was estimated to be 8.94 × 10-4 subs/site/year. Selection pressure analysis showed that two sites 160 and 403 were under positive selection, while E gene is mainly shaped by stronger purifying selection. BSP analysis showed that estimating effective population size from samples of sequences has undergone three obvious increases, additionally, Caribbean and Puerto Rico maintained higher levels of genetic diversity relative to other 6 representative geographical populations using GMRF method. The phylogeographic analysis indicated that two major transmission routes are from South America to Caribbean and East&SouthAsia to Puerto Rico. The trunk reconstruction confirmed that the viral evolution spanned 50 years occurred primarily in Southeast Asia and East&South Asia. In addition, phylogeographic association-trait analysis indicated that the viral phenotypes are highly correlated with phylogeny in Nicaragua and Puerto Rico (P < 0.05).
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Kumar S. R. et al.. Evolution, dispersal and replacement of American genotype dengue type 2 viruses in India (1956–2005): selection pressure and molecular clock analyses. Journal of General Virology 91, 707–720 (2010). - PubMed
-
- Guzman M. G., Alvarez M. & Halstead S. B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Archives of virology 158, 1445–1459 (2013). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

