Gracob: a novel graph-based constant-column biclustering method for mining growth phenotype data
- PMID: 28379298
- PMCID: PMC5870648
- DOI: 10.1093/bioinformatics/btx199
Gracob: a novel graph-based constant-column biclustering method for mining growth phenotype data
Abstract
Motivation: Growth phenotype profiling of genome-wide gene-deletion strains over stress conditions can offer a clear picture that the essentiality of genes depends on environmental conditions. Systematically identifying groups of genes from such high-throughput data that share similar patterns of conditional essentiality and dispensability under various environmental conditions can elucidate how genetic interactions of the growth phenotype are regulated in response to the environment.
Results: We first demonstrate that detecting such 'co-fit' gene groups can be cast as a less well-studied problem in biclustering, i.e. constant-column biclustering. Despite significant advances in biclustering techniques, very few were designed for mining in growth phenotype data. Here, we propose Gracob, a novel, efficient graph-based method that casts and solves the constant-column biclustering problem as a maximal clique finding problem in a multipartite graph. We compared Gracob with a large collection of widely used biclustering methods that cover different types of algorithms designed to detect different types of biclusters. Gracob showed superior performance on finding co-fit genes over all the existing methods on both a variety of synthetic data sets with a wide range of settings, and three real growth phenotype datasets for E. coli, proteobacteria and yeast.
Availability and implementation: Our program is freely available for download at http://sfb.kaust.edu.sa/Pages/Software.aspx.
Contact: xin.gao@kaust.edu.sa.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2017. Published by Oxford University Press.
Figures


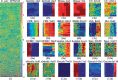
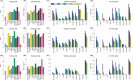
References
-
- Ben-Dor A. et al. (2003) Discovering local structure in gene expression data: the order-preserving submatrix problem. J. Comput. Biol., 10, 373–384. - PubMed
-
- Bergmann S. et al. (2003) Iterative signature algorithm for the analysis of large-scale gene expression data. Phys. Rev. E, 67, 031902. - PubMed
-
- Blattner F.R. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1462. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

