A reference human genome dataset of the BGISEQ-500 sequencer
- PMID: 28379488
- PMCID: PMC5467036
- DOI: 10.1093/gigascience/gix024
A reference human genome dataset of the BGISEQ-500 sequencer
Erratum in
-
Erratum to: A reference human genome dataset of the BGISEQ-500 sequencer.Gigascience. 2018 Dec 1;7(12):giy144. doi: 10.1093/gigascience/giy144. Gigascience. 2018. PMID: 30500904 Free PMC article. No abstract available.
Abstract
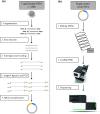
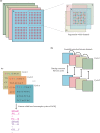
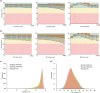
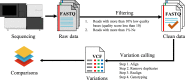
BGISEQ-500 is a new desktop sequencer developed by BGI. Using DNA nanoball and combinational probe anchor synthesis developed from Complete Genomics™ sequencing technologies, it generates short reads at a large scale. Here, we present the first human whole-genome sequencing dataset of BGISEQ-500. The dataset was generated by sequencing the widely used cell line HG001 (NA12878) in two sequencing runs of paired-end 50 bp (PE50) and two sequencing runs of paired-end 100 bp (PE100). We also include examples of the raw images from the sequencer for reference. Finally, we identified variations using this dataset, estimated the accuracy of the variations, and compared to that of the variations identified from similar amounts of publicly available HiSeq2500 data. We found similar single nucleotide polymorphism (SNP) detection accuracy for the BGISEQ-500 PE100 data (false positive rate [FPR] = 0.00020%, sensitivity = 96.20%) compared to the PE150 HiSeq2500 data (FPR = 0.00017%, sensitivity = 96.60%) better SNP detection accuracy than the PE50 data (FPR = 0.0006%, sensitivity = 94.15%). But for insertions and deletions (indels), we found lower accuracy for BGISEQ-500 data (FPR = 0.00069% and 0.00067% for PE100 and PE50 respectively, sensitivity = 88.52% and 70.93%) than the HiSeq2500 data (FPR = 0.00032%, sensitivity = 96.28%). Our dataset can serve as the reference dataset, providing basic information not just for future development, but also for all research and applications based on the new sequencing platform.
Keywords: BGISEQ-500; genomics; next-generation sequencing; sequencing.
© The Authors 2017. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

