Discovery of novel heart rate-associated loci using the Exome Chip
- PMID: 28379579
- PMCID: PMC5458336
- DOI: 10.1093/hmg/ddx113
Discovery of novel heart rate-associated loci using the Exome Chip
Abstract
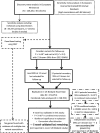
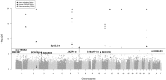
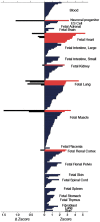
Resting heart rate is a heritable trait, and an increase in heart rate is associated with increased mortality risk. Genome-wide association study analyses have found loci associated with resting heart rate, at the time of our study these loci explained 0.9% of the variation. This study aims to discover new genetic loci associated with heart rate from Exome Chip meta-analyses.Heart rate was measured from either elecrtrocardiograms or pulse recordings. We meta-analysed heart rate association results from 104 452 European-ancestry individuals from 30 cohorts, genotyped using the Exome Chip. Twenty-four variants were selected for follow-up in an independent dataset (UK Biobank, N = 134 251). Conditional and gene-based testing was undertaken, and variants were investigated with bioinformatics methods.We discovered five novel heart rate loci, and one new independent low-frequency non-synonymous variant in an established heart rate locus (KIAA1755). Lead variants in four of the novel loci are non-synonymous variants in the genes C10orf71, DALDR3, TESK2 and SEC31B. The variant at SEC31B is significantly associated with SEC31B expression in heart and tibial nerve tissue. Further candidate genes were detected from long-range regulatory chromatin interactions in heart tissue (SCD, SLF2 and MAPK8). We observed significant enrichment in DNase I hypersensitive sites in fetal heart and lung. Moreover, enrichment was seen for the first time in human neuronal progenitor cells (derived from embryonic stem cells) and fetal muscle samples by including our novel variants.Our findings advance the knowledge of the genetic architecture of heart rate, and indicate new candidate genes for follow-up functional studies.
© The Author 2017. Published by Oxford University Press.
Figures
Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Exploration of haplotype research consortium imputation for genome-wide association studies in 20,032 Generation Scotland participants.Genome Med. 2017 Mar 7;9(1):23. doi: 10.1186/s13073-017-0414-4. Genome Med. 2017. PMID: 28270201 Free PMC article.
-
Discovery and fine-mapping of adiposity loci using high density imputation of genome-wide association studies in individuals of African ancestry: African Ancestry Anthropometry Genetics Consortium.PLoS Genet. 2017 Apr 21;13(4):e1006719. doi: 10.1371/journal.pgen.1006719. eCollection 2017 Apr. PLoS Genet. 2017. PMID: 28430825 Free PMC article.
-
Identification and Functional Assessment of Candidate Causal Cis-Regulatory Variants Underlying Electrocardiographic QT Interval GWAS Loci.Circ Genom Precis Med. 2025 Jun;18(3):e005032. doi: 10.1161/CIRCGEN.124.005032. Epub 2025 May 27. Circ Genom Precis Med. 2025. PMID: 40421528
Cited by
-
Genetic Risk Scores for Complex Disease Traits in Youth.Circ Genom Precis Med. 2020 Aug;13(4):e002775. doi: 10.1161/CIRCGEN.119.002775. Epub 2020 Jun 11. Circ Genom Precis Med. 2020. PMID: 32527150 Free PMC article.
-
Potential functional variants of KIAA genes are associated with breast cancer risk in a case control study.Ann Transl Med. 2021 Apr;9(7):549. doi: 10.21037/atm-20-6108. Ann Transl Med. 2021. PMID: 33987247 Free PMC article.
-
Meta-analysis of up to 622,409 individuals identifies 40 novel smoking behaviour associated genetic loci.Mol Psychiatry. 2020 Oct;25(10):2392-2409. doi: 10.1038/s41380-018-0313-0. Epub 2019 Jan 7. Mol Psychiatry. 2020. PMID: 30617275 Free PMC article.
-
Slowing Heart Rate Protects Against Pathological Cardiac Hypertrophy.Function (Oxf). 2022 Nov 1;4(1):zqac055. doi: 10.1093/function/zqac055. eCollection 2023. Function (Oxf). 2022. PMID: 36540889 Free PMC article.
-
Dissecting mechanisms of chamber-specific cardiac differentiation and its perturbation following retinoic acid exposure.Development. 2022 Jul 1;149(13):dev200557. doi: 10.1242/dev.200557. Epub 2022 Jul 8. Development. 2022. PMID: 35686629 Free PMC article.
References
-
- Aladin A.I., Whelton S.P., Al-Mallah M.H., Blaha M.J., Keteyian S.J., Juraschek S.P., Rubin J., Brawner C.A., Michos E.D. (2014) Relation of resting heart rate to risk for all-cause mortality by gender after considering exercise capacity (the Henry Ford exercise testing project). Am. J. Cardiol., 114, 1701–1706. - PubMed
-
- Carlson N., Dixen U., Marott J.L., Jensen M.T., Jensen G.B. (2014) Predictive value of casual ECG-based resting heart rate compared with resting heart rate obtained from Holter recording. Scand. J. Clin. Lab. Invest., 74, 163–169. - PubMed
-
- Fox K., Bousser M.G., Amarenco P., Chamorro A., Fisher M., Ford I., Hennerici M.G., Mattle H.P., Rothwell P.M. (2013) Heart rate is a prognostic risk factor for myocardial infarction: a post hoc analysis in the PERFORM (Prevention of cerebrovascular and cardiovascular Events of ischemic origin with teRutroban in patients with a history oF ischemic strOke or tRansient ischeMic attack) study population. Int. J. Cardiol., 168, 3500–3505. - PubMed
-
- Woodward M., Webster R., Murakami Y., Barzi F., Lam T.H., Fang X., Suh I., Batty G.D., Huxley R., Rodgers A. (2014) The association between resting heart rate, cardiovascular disease and mortality: evidence from 112,680 men and women in 12 cohorts. Eur. J. Prev. Cardiol., 21, 719–726. - PubMed
-
- Jouven X., Zureik M., Desnos M., Guerot C., Ducimetiere P. (2001) Resting heart rate as a predictive risk factor for sudden death in middle-aged men. Cardiovasc. Res., 50, 373–378. - PubMed
MeSH terms
Grants and funding
- N01 HC095161/HL/NHLBI NIH HHS/United States
- HHSN268201100011I/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- UL1 RR033176/RR/NCRR NIH HHS/United States
- G9521010/MRC_/Medical Research Council/United Kingdom
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- G1001799/MRC_/Medical Research Council/United Kingdom
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- R01 HL103612/HL/NHLBI NIH HHS/United States
- K23 NS059774/NS/NINDS NIH HHS/United States
- HHSN268201100009I/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 HL071251/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- K24 HL105780/HL/NHLBI NIH HHS/United States
- G0000934 /MRC_/Medical Research Council/United Kingdom
- MR/N025083/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201600002C/HL/NHLBI NIH HHS/United States
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
- R01 HL071259/HL/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- HHSN268201000031C/HL/NHLBI NIH HHS/United States
- MR/N01104X/2/MRC_/Medical Research Council/United Kingdom
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- R01 HL071252/HL/NHLBI NIH HHS/United States
- HHSN268201100008I/HL/NHLBI NIH HHS/United States
- HHSN268201100005G/HL/NHLBI NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- RC2 HG005552/HG/NHGRI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- U01 HG006513/HG/NHGRI NIH HHS/United States
- 068545/Z/02/WT_/Wellcome Trust/United Kingdom
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- R01 HL071250/HL/NHLBI NIH HHS/United States
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- MC_PC_14089/MRC_/Medical Research Council/United Kingdom
- R01 HL105756/HL/NHLBI NIH HHS/United States
- MR/L016311/1/MRC_/Medical Research Council/United Kingdom
- MR/N01104X/1/MRC_/Medical Research Council/United Kingdom
- MR/N005813/1/MRC_/Medical Research Council/United Kingdom
- P30 DK063491/DK/NIDDK NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- HHSN268201300049C/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- R01 NS059727/NS/NINDS NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- HHSN268201600003C/HL/NHLBI NIH HHS/United States
- HHSN268201100005I/HL/NHLBI NIH HHS/United States
- R01 HG007089/HG/NHGRI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- HHSN268201300047C/HL/NHLBI NIH HHS/United States
- HHSN271201100005C/DA/NIDA NIH HHS/United States
- HHSN268201300050C/HL/NHLBI NIH HHS/United States
- HHSN268201600004C/HL/NHLBI NIH HHS/United States
- MC_PC_U127561128/MRC_/Medical Research Council/United Kingdom
- MR/M501633/2/MRC_/Medical Research Council/United Kingdom
- N01 HC095163/HL/NHLBI NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- HHSN268201600001C/HL/NHLBI NIH HHS/United States
- MR/K006584/1/MRC_/Medical Research Council/United Kingdom
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- R01 NS073344/NS/NINDS NIH HHS/United States
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- R01 HL071205/HL/NHLBI NIH HHS/United States
- S10 OD020069/OD/NIH HHS/United States
- HHSN268201100007I/HL/NHLBI NIH HHS/United States
- RG/14/5/30893/BHF_/British Heart Foundation/United Kingdom
- MR/K007017/1/MRC_/Medical Research Council/United Kingdom
- UL1 TR000040/TR/NCATS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- HHSN268201300046C/HL/NHLBI NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- R01 HG007022/HG/NHGRI NIH HHS/United States
- U01 HL117626/HL/NHLBI NIH HHS/United States
- MC_PC_13040/MRC_/Medical Research Council/United Kingdom
- RC2 HG005581/HG/NHGRI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- PG/17/59/33139/BHF_/British Heart Foundation/United Kingdom
- N01 HC095165/HL/NHLBI NIH HHS/United States
- G0600237/MRC_/Medical Research Council/United Kingdom
- N01 HC095164/HL/NHLBI NIH HHS/United States
- R01 HL071258/HL/NHLBI NIH HHS/United States
- K23 HL114724/HL/NHLBI NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- R01 HL111089/HL/NHLBI NIH HHS/United States
- R01 HL116747/HL/NHLBI NIH HHS/United States
- PG/12/38/29615/BHF_/British Heart Foundation/United Kingdom
- MR/M501633/1/MRC_/Medical Research Council/United Kingdom
- N01 HC095160/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

