Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates
- PMID: 28379941
- PMCID: PMC11348803
- DOI: 10.1038/nature21722
Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates
Erratum in
-
Author Correction: Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates.Nature. 2024 Jun;630(8017):E11. doi: 10.1038/s41586-024-07594-8. Nature. 2024. PMID: 38834754 No abstract available.
Abstract
Normal differentiation and induced reprogramming require the activation of target cell programs and silencing of donor cell programs. In reprogramming, the same factors are often used to reprogram many different donor cell types. As most developmental repressors, such as RE1-silencing transcription factor (REST) and Groucho (also known as TLE), are considered lineage-specific repressors, it remains unclear how identical combinations of transcription factors can silence so many different donor programs. Distinct lineage repressors would have to be induced in different donor cell types. Here, by studying the reprogramming of mouse fibroblasts to neurons, we found that the pan neuron-specific transcription factor Myt1-like (Myt1l) exerts its pro-neuronal function by direct repression of many different somatic lineage programs except the neuronal program. The repressive function of Myt1l is mediated via recruitment of a complex containing Sin3b by binding to a previously uncharacterized N-terminal domain. In agreement with its repressive function, the genomic binding sites of Myt1l are similar in neurons and fibroblasts and are preferentially in an open chromatin configuration. The Notch signalling pathway is repressed by Myt1l through silencing of several members, including Hes1. Acute knockdown of Myt1l in the developing mouse brain mimicked a Notch gain-of-function phenotype, suggesting that Myt1l allows newborn neurons to escape Notch activation during normal development. Depletion of Myt1l in primary postmitotic neurons de-repressed non-neuronal programs and impaired neuronal gene expression and function, indicating that many somatic lineage programs are actively and persistently repressed by Myt1l to maintain neuronal identity. It is now tempting to speculate that similar 'many-but-one' lineage repressors exist for other cell fates; such repressors, in combination with lineage-specific activators, would be prime candidates for use in reprogramming additional cell types.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

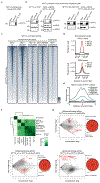







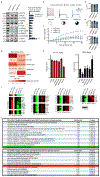




Comment in
-
Neural development: Keeping a lid on alternative fates.Nat Rev Neurosci. 2017 Jun;18(6):323. doi: 10.1038/nrn.2017.54. Epub 2017 Apr 27. Nat Rev Neurosci. 2017. PMID: 28446787 No abstract available.
References
-
- Schäfer BW, Blakely BT, Darlington GJ & Blau HM Effect of cell history on response to helix-loop-helix family of myogenic regulators. Nature 344, 454–458 (1990). - PubMed
-
- Terranova R, Pereira CF, Roure, Du, C., Merkenschlager, M. & Fisher, A. G. Acquisition and extinction of gene expression programs are separable events in heterokaryon reprogramming. J Cell Sci 119, 2065–2072 (2006). - PubMed
-
- Xu J, Du Y & Deng H Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell 16, 119–134 (2015). - PubMed
-
- Schoenherr CJ & Anderson DJ The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363 (1995). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

