IL-15 regulates migration, invasion, angiogenesis and genes associated with lipid metabolism and inflammation in prostate cancer
- PMID: 28379958
- PMCID: PMC5381801
- DOI: 10.1371/journal.pone.0172786
IL-15 regulates migration, invasion, angiogenesis and genes associated with lipid metabolism and inflammation in prostate cancer
Abstract
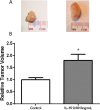
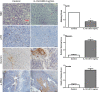
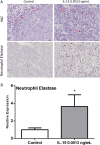
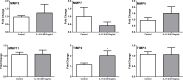
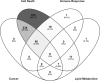
Prostate cancer (PCa) is the most commonly diagnosed non-cutaneous cancer. In the United States it is second leading cause of cancer related deaths in men. PCa is often treated via radical prostatectomy (RP). However, 15-30% of the patients develop biochemical recurrence (i.e. increased serum prostate specific antigen (PSA) levels). Interleukin-15 (IL-15) is a secreted cytokine found over expressed in patients with recurrence-free survival after RP. In our study, we aim to determine the role of IL-15 in PCa using in vitro and in vivo models, and gene expression analysis. PC3 (androgen-independent) and 22RV1 (androgen-dependent) cell lines were treated with IL-15 at 0.0013 ng/mL and 0.1 ng/mL. Tumor growth was evaluated using an orthotopic xenograft model. The anterior prostate lobes of SCID mice were injected with 250,000 22RV1 cells and IL-15 was administered bi-weekly with intraperitoneal (IP) injections during 4 weeks. Tumor tissue was collected for immunohistochemical and gene expression analysis. To study changes in gene expression, we looked at "Tumor Metastasis" and "PI3K pathway" using commercially available PCR arrays. In addition, we employed a microarray approach using the Affymetrix Hugene 2.0 ST array chip followed by analysis with Ingenuity Pathways Analysis (IPA) software. In vitro studies showed that IL-15 decreased PCa cell motility at both concentrations. In vivo studies showed that IL-15 increased neutrophil infiltration, and the expression of adiponectin, desmin and alpha smooth muscle actin (α-sma) in the tumor tissue. Angiogenesis analysis, using CD31 immunohistochemistry, showed that IL-15 decreased the number of blood vessels. Gene expression analysis identified Cancer, Cell Death, Immune Response and Lipid Metabolism as the major diseases and functions altered in tumors treated with IL-15. This suggests that IL-15 causes inflammation and changes in stroma that can promote decreased tumor cell proliferation.
Conflict of interest statement
Figures








References
-
- Bruce JY, Lang JM, McNeel DG, Liu G. Current controversies in the management of biochemical failure in prostate cancer. Clin Adv Hematol Oncol. 2012;10: 716–22. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

