SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion
- PMID: 28380376
- PMCID: PMC5444661
- DOI: 10.1016/j.cmet.2017.03.003
SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion
Abstract
Sirtuins are NAD+-dependent protein deacylases that regulate several aspects of metabolism and aging. In contrast to the other mammalian sirtuins, the primary enzymatic activity of mitochondrial sirtuin 4 (SIRT4) and its overall role in metabolic control have remained enigmatic. Using a combination of phylogenetics, structural biology, and enzymology, we show that SIRT4 removes three acyl moieties from lysine residues: methylglutaryl (MG)-, hydroxymethylglutaryl (HMG)-, and 3-methylglutaconyl (MGc)-lysine. The metabolites leading to these post-translational modifications are intermediates in leucine oxidation, and we show a primary role for SIRT4 in controlling this pathway in mice. Furthermore, we find that dysregulated leucine metabolism in SIRT4KO mice leads to elevated basal and stimulated insulin secretion, which progressively develops into glucose intolerance and insulin resistance. These findings identify a robust enzymatic activity for SIRT4, uncover a mechanism controlling branched-chain amino acid flux, and position SIRT4 as a crucial player maintaining insulin secretion and glucose homeostasis during aging.
Keywords: branched-chain amino acids; deacylase; insulin secretion; leucine; mitochondria; sirtuin 4.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


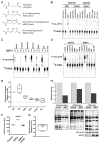


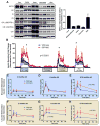
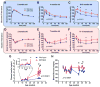
Comment in
-
For Certain, SIRT4 Activities!Trends Biochem Sci. 2017 Jul;42(7):499-501. doi: 10.1016/j.tibs.2017.05.008. Epub 2017 Jun 3. Trends Biochem Sci. 2017. PMID: 28587732 Free PMC article.
References
-
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. - PubMed
-
- Allen A, Kwagh J, Fang J, Stanley CA, Smith TJ. Evolution of glutamate dehydrogenase regulation of insulin homeostasis is an example of molecular exaptation. Biochemistry. 2004;43:14431–14443. - PubMed
-
- Campbell JE, Ussher JR, Mulvihill EE, Kolic J, Baggio LL, Cao X, Liu Y, Lamont BJ, Morii T, Streutker CJ, et al. TCF1 links GIPR signaling to the control of beta cell function and survival. Nat Med. 2016;22:84–90. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

