Transcriptional signatures of steroid hormones in the striatal neurons and astrocytes
- PMID: 28381250
- PMCID: PMC5381047
- DOI: 10.1186/s12868-017-0352-5
Transcriptional signatures of steroid hormones in the striatal neurons and astrocytes
Abstract
Background: The mechanisms of steroids actions in the brain mainly involve the binding and nuclear translocation of specific cytoplasmic receptors. These receptors can act as transcription factors and regulate gene expression. However, steroid-dependent transcriptional regulation in different types of neural cells is not yet fully understood. The aim of this study was to evaluate and compare transcriptional alterations induced by various steroid receptor agonists in primary cultures of astrocytes and neurons from mouse brain.
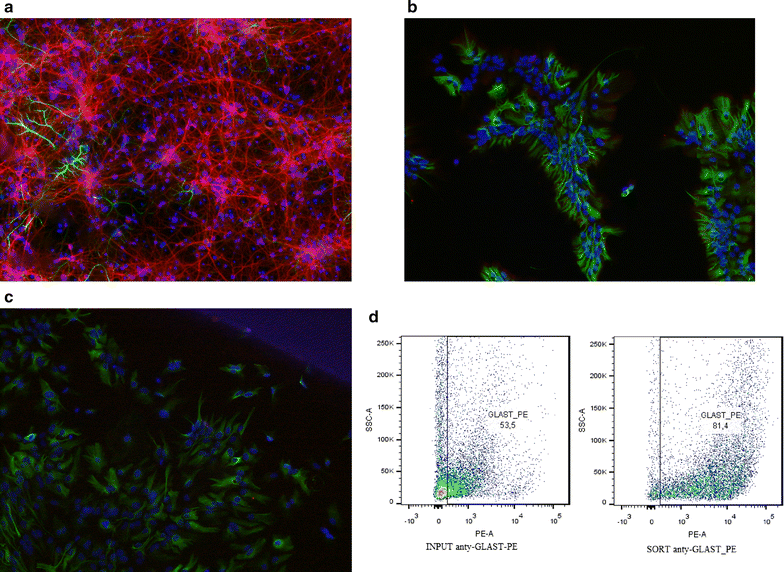
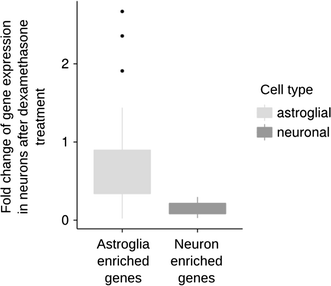
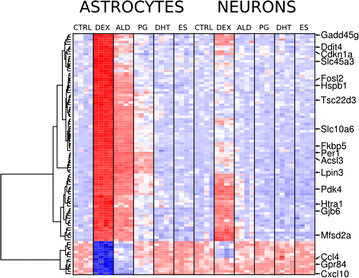
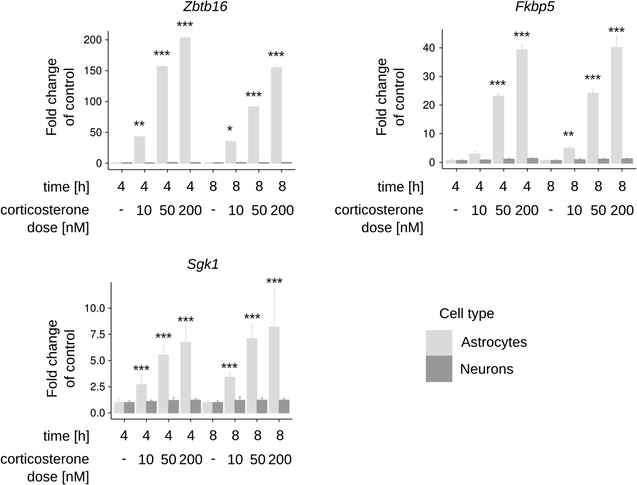
Results: We utilized whole-genome microarrays (Illumina Mouse WG-6) and quantitative PCR analyses to measure mRNA abundance levels. To stimulate gene expression we treated neuronal and astroglial cultures with dexamethasone (100 nM), aldosterone (200 nM), progesterone (200 nM), 5α-dihydrotestosterone (200 nM) and β-Estradiol (200 nM) for 4 h. Neurons were found to exhibit higher levels of expression of mineralocorticoid receptor, progesterone receptor and estrogen receptor 2 than astrocytes. However, higher mRNA level of glucocorticoid receptor mRNA was observed in astrocytes. We identified 956 genes regulated by steroids. In astrocytes we found 381 genes altered by dexamethasone and 19 altered by aldosterone. Functional classification of the regulated genes indicated their putative involvement in multiple aspects of cell metabolism (up-regulated Slc2a1, Pdk4 and Slc45a3) and the inflammatory response (down-regulated Ccl3, Il1b and Tnf). Progesterone, dihydrotestosterone and estradiol did not change gene expression in astrocytes. We found no significant changes in gene expression in neurons.
Conclusions: The obtained results indicate that glial cells might be the primary targets of transcriptional action of steroids in the central nervous system. Substantial changes in gene expression driven by the glucocorticoid receptor imply an important role for the hypothalamic-pituitary-adrenal axis in the hormone-dependent regulation of brain physiology. This is an in vitro study. Hence, the model may not accurately reflect all the effects of steroids on gene expression in neurons in vivo.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

