Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix
- PMID: 28381423
- PMCID: PMC5449147
- DOI: 10.1091/mbc.E16-09-0654
Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix
Abstract
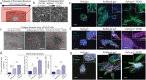
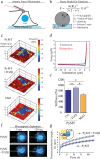
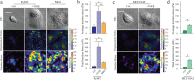
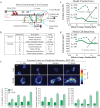
Metastasis requires tumor cells to navigate through a stiff stroma and squeeze through confined microenvironments. Whether tumors exploit unique biophysical properties to metastasize remains unclear. Data show that invading mammary tumor cells, when cultured in a stiffened three-dimensional extracellular matrix that recapitulates the primary tumor stroma, adopt a basal-like phenotype. Metastatic tumor cells and basal-like tumor cells exert higher integrin-mediated traction forces at the bulk and molecular levels, consistent with a motor-clutch model in which motors and clutches are both increased. Basal-like nonmalignant mammary epithelial cells also display an altered integrin adhesion molecular organization at the nanoscale and recruit a suite of paxillin-associated proteins implicated in invasion and metastasis. Phosphorylation of paxillin by Src family kinases, which regulates adhesion turnover, is similarly enhanced in the metastatic and basal-like tumor cells, fostered by a stiff matrix, and critical for tumor cell invasion in our assays. Bioinformatics reveals an unappreciated relationship between Src kinases, paxillin, and survival of breast cancer patients. Thus adoption of the basal-like adhesion phenotype may favor the recruitment of molecules that facilitate tumor metastasis to integrin-based adhesions. Analysis of the physical properties of tumor cells and integrin adhesion composition in biopsies may be predictive of patient outcome.
© 2017 Mekhdjian, Kai, Rubashkin, et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

