SETD1B Activates iNOS Expression in Myeloid-Derived Suppressor Cells
- PMID: 28381543
- PMCID: PMC5495112
- DOI: 10.1158/0008-5472.CAN-16-2238
SETD1B Activates iNOS Expression in Myeloid-Derived Suppressor Cells
Abstract
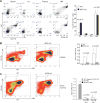
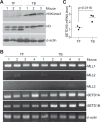
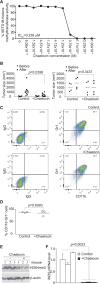
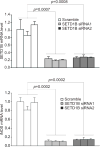
Inducible nitric oxide synthase (iNOS) generates nitric oxide (NO) in myeloid cells that acts as a defense mechanism to suppress invading microorganisms or neoplastic cells. In tumor-bearing mice, elevated iNOS expression is a hallmark of myeloid-derived suppressor cells (MDSC). MDSCs use NO to nitrate both the T-cell receptor and STAT1, thus inhibiting T-cell activation and the antitumor immune response. The molecular mechanisms underlying iNOS expression and regulation in tumor-induced MDSCs are unknown. We report here that deficiency in IRF8 results in diminished iNOS expression in both mature CD11b+Gr1- and immature CD11b+Gr1+ myeloid cells in vivo Strikingly, although IRF8 was silenced in tumor-induced MDSCs, iNOS expression was significantly elevated in tumor-induced MDSCs, suggesting that the expression of iNOS is regulated by an IRF8-independent mechanism under pathologic conditions. Furthermore, tumor-induced MDSCs exhibited diminished STAT1 and NF-κB Rel protein levels, the essential inducers of iNOS in myeloid cells. Instead, tumor-induced MDSCs showed increased SETD1B expression as compared with their cellular equivalents in tumor-free mice. Chromatin immunoprecipitation revealed that H3K4me3, the target of SETD1B, was enriched at the nos2 promoter in tumor-induced MDSCs, and inhibition or silencing of SETD1B diminished iNOS expression in tumor-induced MDSCs. Our results show how tumor cells use the SETD1B-H3K4me3 epigenetic axis to bypass a normal role for IRF8 expression in activating iNOS expression in MDSCs when they are generated under pathologic conditions. Cancer Res; 77(11); 2834-43. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
Conflict of interest: The authors declare no potential conflicts of interest
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

