The chromatin-remodeling subunit Baf200 promotes homology-directed DNA repair and regulates distinct chromatin-remodeling complexes
- PMID: 28381560
- PMCID: PMC5437250
- DOI: 10.1074/jbc.M117.778183
The chromatin-remodeling subunit Baf200 promotes homology-directed DNA repair and regulates distinct chromatin-remodeling complexes
Abstract
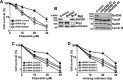
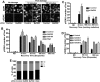
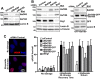
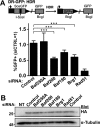
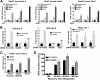
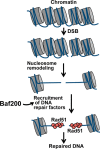
The efficiency and type of pathway chosen to repair DNA double-strand breaks (DSBs) are critically influenced by the nucleosome packaging and the chromatin architecture surrounding the DSBs. The Swi/Snf (PBAF and BAF) chromatin-remodeling complexes contribute to DNA damage-induced nucleosome remodeling, but the mechanism by which it contributes to this function is poorly understood. Herein, we report how the Baf200 (Arid2) PBAF-defining subunit regulates DSB repair. We used cytological and biochemical approaches to show that Baf200 plays an important function by facilitating homologous recombination-dependent processes, such as recruitment of Rad51 (a key component of homologous recombination) to DSBs, homology-directed repair, and cell survival after DNA damage. Furthermore, we observed that Baf200 and Rad51 are present in the same complex and that this interaction is mediated by C-terminal sequences in both proteins. It has been recognized previously that the interplay between distinct forms of Swi/Snf has profound functional consequences, but we understand little about the composition of complexes formed by PBAF protein subunits. Our biochemical analyses reveal that Baf200 forms at least two distinct complexes. One is a canonical form of PBAF including the Swi/Snf-associated Brg1 catalytic subunit, and the other contains Baf180 but not Brg1. This distinction of PBAF complexes based on their unique composition provides the foundation for future studies on the specific contributions of the PBAF forms to the regulation of DNA repair.
Keywords: Baf200; DNA damage; DNA recombination; DNA repair; PBAF; Rad51; chromatin remodeling; homologous recombination.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the content of this article
Figures



References
-
- Jeggo P. A., and Downs J. A. (2014) Roles of chromatin remodellers in DNA double strand break repair. Exp. Cell Res. 329, 69–77 - PubMed
-
- Seeber A., Hauer M., and Gasser S. M. (2013) Nucleosome remodelers in double-strand break repair. Curr. Opin. Genet. Dev. 23, 174–184 - PubMed
-
- Goodarzi A. A., Jeggo P., and Lobrich M. (2010) The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Repair 9, 1273–1282 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

