Understanding the Mechanism of the Broad-Spectrum Antiviral Activity of Favipiravir (T-705): Key Role of the F1 Motif of the Viral Polymerase
- PMID: 28381577
- PMCID: PMC5446660
- DOI: 10.1128/JVI.00487-17
Understanding the Mechanism of the Broad-Spectrum Antiviral Activity of Favipiravir (T-705): Key Role of the F1 Motif of the Viral Polymerase
Abstract
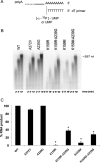
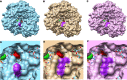
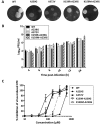
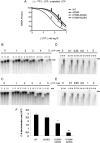
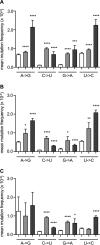
Favipiravir (T-705) is a broad-spectrum antiviral agent that has been approved in Japan for the treatment of influenza virus infections. T-705 also inhibits the replication of various RNA viruses, including chikungunya virus (CHIKV). We demonstrated earlier that the K291R mutation in the F1 motif of the RNA-dependent RNA polymerase (RdRp) of CHIKV is responsible for low-level resistance to T-705. Interestingly, this lysine is highly conserved in the RdRp of positive-sense single-stranded RNA (+ssRNA) viruses. To obtain insights into the unique broad-spectrum antiviral activity of T-705, we explored the role of this lysine using another +ssRNA virus, namely, coxsackievirus B3 (CVB3). Introduction of the corresponding K-to-R substitution in the CVB3 RdRp (K159R) resulted in a nonviable virus. Replication competence of the K159R variant was restored by spontaneous acquisition of an A239G substitution in the RdRp. A mutagenesis analysis at position K159 identified the K159M variant as the only other viable variant which had also acquired the A239G substitution. The K159 substitutions markedly decreased the processivity of the purified viral RdRp, which was restored by the introduction of the A239G mutation. The K159R A239G and K159M A239G variants proved, surprisingly, more susceptible than the wild-type virus to T-705 and exhibited lower fidelity in polymerase assays. Furthermore, the K159R A239G variant was found to be highly attenuated in mice. We thus demonstrate that the conserved lysine in the F1 motif of the RdRp of +ssRNA viruses is involved in the broad-spectrum antiviral activity of T-705 and that it is a key amino acid for the proper functioning of the enzyme.IMPORTANCE In this study, we report the key role of a highly conserved lysine residue of the viral polymerase in the broad-spectrum antiviral activity of favipiravir (T-705) against positive-sense single-stranded RNA viruses. Substitutions of this conserved lysine have a major negative impact on the functionality of the RdRp. Furthermore, we show that this lysine is involved in the fidelity of the RdRp and that the RdRp fidelity influences the sensitivity of the virus for the antiviral efficacy of T-705. Consequently, these results provide insights into the mechanism of the antiviral activity of T-705 and may lay the basis for the design of novel chemical scaffolds that may be endowed with a more potent broad-spectrum antiviral activity than that of T-705.
Keywords: CVB3; RdRp; favipiravir; fidelity; mutagenesis.
Copyright © 2017 American Society for Microbiology.
Figures

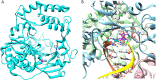


References
-
- Zmurko J, Marques RE, Schols D, Verbeken E, Kaptein SJF, Neyts J. 2016. The viral polymerase inhibitor 7-deaza-2′-C-methyladenosine is a potent inhibitor of in vitro zika virus replication and delays disease progression in a robust mouse infection model. PLoS Negl Trop Dis 10:e0004695. doi:10.1371/journal.pntd.0004695. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

