Identifying genes for neurobehavioural traits in rodents: progress and pitfalls
- PMID: 28381599
- PMCID: PMC5399566
- DOI: 10.1242/dmm.027789
Identifying genes for neurobehavioural traits in rodents: progress and pitfalls
Abstract
Identifying genes and pathways that contribute to differences in neurobehavioural traits is a key goal in psychiatric research. Despite considerable success in identifying quantitative trait loci (QTLs) associated with behaviour in laboratory rodents, pinpointing the causal variants and genes is more challenging. For a long time, the main obstacle was the size of QTLs, which could encompass tens if not hundreds of genes. However, recent studies have exploited mouse and rat resources that allow mapping of phenotypes to narrow intervals, encompassing only a few genes. Here, we review these studies, showcase the rodent resources they have used and highlight the insights into neurobehavioural traits provided to date. We discuss what we see as the biggest challenge in the field - translating QTLs into biological knowledge by experimentally validating and functionally characterizing candidate genes - and propose that the CRISPR/Cas genome-editing system holds the key to overcoming this obstacle. Finally, we challenge traditional views on inbred versus outbred resources in the light of recent resource and technology developments.
Keywords: Genetics of behaviour; Quantitative trait loci; Rodent resources.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures
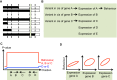
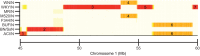
References
-
- Allen H. L., Estrada K., Lettre G., Berndt S. I., Weedon M. N., Rivadeneira F., Willer C. J., Jackson A. U., Vedantam S., Raychaudhuri S. et al. (2010). Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832-838. 10.1038/nature09410 - DOI - PMC - PubMed
-
- American Psychiatric Association (2000). Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition: DSM-IV-TR®. Washington, DC: American Psychiatric Association.
-
- Andreux P. A., Williams E. G., Koutnikova H., Houtkooper R. H., Champy M.-F., Henry H., Schoonjans K., Williams R. W. and Auwerx J. (2012). Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell 150, 1287-1299. 10.1016/j.cell.2012.08.012 - DOI - PMC - PubMed
-
- Atanur S. S., Diaz A. G., Maratou K., Sarkis A., Rotival M., Game L., Tschannen M. R., Kaisaki P. J., Otto G. W., Ma M. C. J. et al. (2013). Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell 154, 691-703. 10.1016/j.cell.2013.06.040 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

