The developmental proteome of Drosophila melanogaster
- PMID: 28381612
- PMCID: PMC5495078
- DOI: 10.1101/gr.213694.116
The developmental proteome of Drosophila melanogaster
Abstract
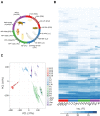
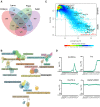
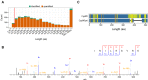
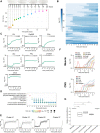
Drosophila melanogaster is a widely used genetic model organism in developmental biology. While this model organism has been intensively studied at the RNA level, a comprehensive proteomic study covering the complete life cycle is still missing. Here, we apply label-free quantitative proteomics to explore proteome remodeling across Drosophila's life cycle, resulting in 7952 proteins, and provide a high temporal-resolved embryogenesis proteome of 5458 proteins. Our proteome data enabled us to monitor isoform-specific expression of 34 genes during development, to identify the pseudogene Cyp9f3Ψ as a protein-coding gene, and to obtain evidence of 268 small proteins. Moreover, the comparison with available transcriptomic data uncovered examples of poor correlation between mRNA and protein, underscoring the importance of proteomics to study developmental progression. Data integration of our embryogenesis proteome with tissue-specific data revealed spatial and temporal information for further functional studies of yet uncharacterized proteins. Overall, our high resolution proteomes provide a powerful resource and can be explored in detail in our interactive web interface.
© 2017 Casas-Vila et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. - PubMed
-
- Bonaldi T, Straub T, Cox J, Kumar C, Becker PB, Mann M. 2008. Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol Cell 31: 762–772. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
