Genetic variations in cancer-related significantly mutated genes and lung cancer susceptibility
- PMID: 28383694
- PMCID: PMC5834153
- DOI: 10.1093/annonc/mdx161
Genetic variations in cancer-related significantly mutated genes and lung cancer susceptibility
Abstract
Background: Cancer initiation and development are driven by key mutations in driver genes. Applying high-throughput sequencing technologies and bioinformatic analyses, The Cancer Genome Atlas (TCGA) project has identified panels of somatic mutations that contributed to the etiology of various cancers. However, there are few studies investigating the germline genetic variations in these significantly mutated genes (SMGs) and lung cancer susceptibility.
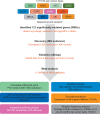
Patients and methods: We comprehensively evaluated 1655 tagged single nucleotide polymorphisms (SNPs) located in 127 SMGs identified by TCGA, and test their association with lung cancer risk in large-scale case-control study. Functional effect of the validated SNPs, gene mutation frequency and pathways were analyzed.
Results: We found 11 SNPs in 8 genes showed consistent association (P < 0.1) and 8 SNPs significantly associated with lung cancer risk (P < 0.05) in both discovery and validation phases. The most significant association was rs10412613 in PPP2R1A, with the minor G allele associated with a decreased risk of lung cancer [odds ratio = 0.91, 95% confidence interval (CI): 0.87-0.96, P = 2.3 × 10-4]. Cumulative analysis of risk score built as a weight sum of the 11 SNPs showed consistently elevated risk with increasing risk score (P for trend = 9.5 × 10-9). In stratified analyses, the association of PPP2R1A:rs10412613 and lung cancer risk appeared stronger among population of younger age at diagnosis and never smokers. The expression quantitative trait loci analysis indicated that rs10412613, rs10804682, rs635469 and rs6742399 genotypes significantly correlated with the expression of PPP2R1A, ATR, SETBP1 and ERBB4, respectively. From TCGA data, expression of the identified genes was significantly different in lung tumors compared with normal tissues, and the genes' highest mutation frequency was found in lung cancers. Integrative pathway analysis indicated the identified genes were mainly involved in AKT/NF-κB regulatory pathway suggesting the underlying biological processes.
Conclusion: This study revealed novel genetic variants in SMGs associated with lung cancer risk, which might contribute to elucidating the biological network involved in lung cancer development.
Keywords: TCGA; eQTL; lung cancer; pathway analysis; significantly mutated genes; tagSNP.
© The Author 2017. Published by Oxford University Press on behalf of the European Society for Medical Oncology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2015. CA Cancer J Clin 2015; 65: 5–29. - PubMed
-
- Ruano-Ravina A, Figueiras A, Barros-Dios JM.. Lung cancer and related risk factors: an update of the literature. Public Health 2003; 117: 149–156. - PubMed
-
- Sato M, Shames DS, Gazdar AF, Minna JD.. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol 2007; 2: 327–343. - PubMed
-
- Meyerson M, Franklin WA, Kelley MJ.. Molecular classification and molecular genetics of human lung cancers. Semin Oncol 2004; 31: 4–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

