Scarless assembly of unphosphorylated DNA fragments with a simplified DATEL method
- PMID: 28384080
- PMCID: PMC5470510
- DOI: 10.1080/21655979.2017.1308986
Scarless assembly of unphosphorylated DNA fragments with a simplified DATEL method
Abstract
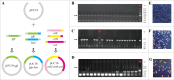
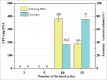
Efficient assembly of multiple DNA fragments is a pivotal technology for synthetic biology. A scarless and sequence-independent DNA assembly method (DATEL) using thermal exonucleases has been developed recently. Here, we present a simplified DATEL (sDATEL) for efficient assembly of unphosphorylated DNA fragments with low cost. The sDATEL method is only dependent on Taq DNA polymerase and Taq DNA ligase. After optimizing the committed parameters of the reaction system such as pH and the concentration of Mg2+ and NAD+, the assembly efficiency was increased by 32-fold. To further improve the assembly capacity, the number of thermal cycles was optimized, resulting in successful assembly 4 unphosphorylated DNA fragments with an accuracy of 75%. sDATEL could be a desirable method for routine manual and automated assembly.
Keywords: DNA assembly; PCR; Simplified DATEL; Synthetic biology; Unphosphorylated DNA fragments.
Figures

Similar articles
-
DATEL: A Scarless and Sequence-Independent DNA Assembly Method Using Thermostable Exonucleases and Ligase.ACS Synth Biol. 2016 Sep 16;5(9):1028-32. doi: 10.1021/acssynbio.6b00078. Epub 2016 Jun 1. ACS Synth Biol. 2016. PMID: 27230689
-
DNA Assembly with the DATEL Method.Methods Mol Biol. 2018;1772:421-428. doi: 10.1007/978-1-4939-7795-6_24. Methods Mol Biol. 2018. PMID: 29754243
-
Enzymatic assembly of overlapping DNA fragments.Methods Enzymol. 2011;498:349-61. doi: 10.1016/B978-0-12-385120-8.00015-2. Methods Enzymol. 2011. PMID: 21601685 Free PMC article.
-
[Perspective on the novel methods for DNA assembly].Sheng Wu Gong Cheng Xue Bao. 2013 Aug;29(8):1113-22. Sheng Wu Gong Cheng Xue Bao. 2013. PMID: 24364348 Review. Chinese.
-
High molecular weight DNA assembly in vivo for synthetic biology applications.Crit Rev Biotechnol. 2017 May;37(3):277-286. doi: 10.3109/07388551.2016.1141394. Epub 2016 Feb 10. Crit Rev Biotechnol. 2017. PMID: 26863154 Review.
Cited by
-
The evolving art of creating genetic diversity: From directed evolution to synthetic biology.Biotechnol Adv. 2021 Sep-Oct;50:107762. doi: 10.1016/j.biotechadv.2021.107762. Epub 2021 May 15. Biotechnol Adv. 2021. PMID: 34000294 Free PMC article. Review.
References
-
- Ellis T, Adie T, Baldwin GS. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr Biol-UK 2011; 3:109-18; PMID:21246151; https://doi.org/10.1039/c0ib00070a - DOI - PubMed
-
- Kang Z, Zhang J, Jin P, Yang S. Directed evolution combined with synthetic biology strategies expedite semi-rational engineering of genes and genomes. Bioengineered 2015; 6:136-40; PMID:25621864; https://doi.org/10.4161/bioe.34347 - DOI - PMC - PubMed
-
- Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci U S A 2004; 101:15573-8; PMID:15496466; https://doi.org/10.1073/pnas.0406911101 - DOI - PMC - PubMed
-
- Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 2009; 4:e5553; PMID:19436741; https://doi.org/10.1371/journal.pone.0005553 - DOI - PMC - PubMed
-
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juarez P, Fernandez-del-Carmen A, Granell A, Orzaez D. GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PLoS One 2011; 6:e21622; PMID:21750718; https://doi.org/10.1371/journal.pone.0021622 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
