Analyses of the probiotic property and stress resistance-related genes of Lactococcus lactis subsp. lactis NCDO 2118 through comparative genomics and in vitro assays
- PMID: 28384209
- PMCID: PMC5383145
- DOI: 10.1371/journal.pone.0175116
Analyses of the probiotic property and stress resistance-related genes of Lactococcus lactis subsp. lactis NCDO 2118 through comparative genomics and in vitro assays
Abstract
Lactococcus lactis subsp. lactis NCDO 2118 was recently reported to alleviate colitis symptoms via its anti-inflammatory and immunomodulatory activities, which are exerted by exported proteins that are not produced by L. lactis subsp. lactis IL1403. Here, we used in vitro and in silico approaches to characterize the genomic structure, the safety aspects, and the immunomodulatory activity of this strain. Through comparative genomics, we identified genomic islands, phage regions, bile salt and acid stress resistance genes, bacteriocins, adhesion-related and antibiotic resistance genes, and genes encoding proteins that are putatively secreted, expressed in vitro and absent from IL1403. The high degree of similarity between all Lactococcus suggests that the Symbiotic Islands commonly shared by both NCDO 2118 and KF147 may be responsible for their close relationship and their adaptation to plants. The predicted bacteriocins may play an important role against the invasion of competing strains. The genes related to the acid and bile salt stresses may play important roles in gastrointestinal tract survival, whereas the adhesion proteins are important for persistence in the gut, culminating in the competitive exclusion of other bacteria. Finally, the five secreted and expressed proteins may be important targets for studies of new anti-inflammatory and immunomodulatory proteins. Altogether, the analyses performed here highlight the potential use of this strain as a target for the future development of probiotic foods.
Conflict of interest statement
Figures

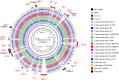

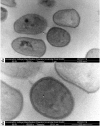
References
-
- Khalid K. An overview of lactic acid bacteria. International Journal of Biosciences. 2011;1: 1–13.
-
- Pfeiler EA, Klaenhammer TR. The genomics of lactic acid bacteria. Trends Microbiol. 2007;15: 546–553. doi: 10.1016/j.tim.2007.09.010 - DOI - PubMed
-
- Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, et al. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001;11: 731–753. doi: 10.1101/gr.169701 - DOI - PMC - PubMed
-
- Smeianov V, Wechter P, Broadbent J, Hughes J, Rodríguez B, Christensen TK, et al. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 2007;73: 2661–2672. doi: 10.1128/AEM.00005-07 - DOI - PMC - PubMed
-
- Díaz-Muñiz I, Banavara DS, Budinich MF, Rankin SA, Dudley EG, Steele JL, et al. Lactobacillus casei metabolic potential to utilize citrate as an energy source in ripening cheese: a bioinformatics approach. J Appl Microbiol. 2006;101: 872–882. doi: 10.1111/j.1365-2672.2006.02965.x - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

