Transmembrane domain quality control systems operate at the endoplasmic reticulum and Golgi apparatus
- PMID: 28384259
- PMCID: PMC5383021
- DOI: 10.1371/journal.pone.0173924
Transmembrane domain quality control systems operate at the endoplasmic reticulum and Golgi apparatus
Abstract
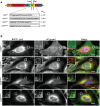
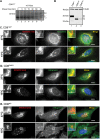
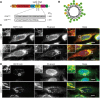
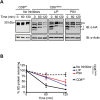
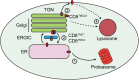
Multiple protein quality control systems operate to ensure that misfolded proteins are efficiently cleared from the cell. While quality control systems that assess the folding status of soluble domains have been extensively studied, transmembrane domain (TMD) quality control mechanisms are poorly understood. Here, we have used chimeras based on the type I plasma membrane protein CD8 in which the endogenous TMD was substituted with transmembrane sequences derived from different polytopic membrane proteins as a mode to investigate the quality control of unassembled TMDs along the secretory pathway. We find that the three TMDs examined prevent trafficking of CD8 to the cell surface via potentially distinct mechanisms. CD8 containing two distinct non-native transmembrane sequences escape the ER and are subsequently retrieved from the Golgi, possibly via Rer1, leading to ER localisation at steady state. A third chimera, containing an altered transmembrane domain, was predominantly localised to the Golgi at steady state, indicating the existence of an additional quality control checkpoint that identifies non-native transmembrane domains that have escaped ER retention and retrieval. Preliminary experiments indicate that protein retained by quality control mechanisms at the Golgi are targeted to lysosomes for degradation.
Conflict of interest statement
Figures


References
-
- Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7(4):1029–38. Epub 1998/05/06. PubMed Central PMCID: PMCPmc2143985. doi: 10.1002/pro.5560070420 - DOI - PMC - PubMed
-
- Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5(5):a013201 Epub 2013/05/03. doi: 10.1101/cshperspect.a013201 - DOI - PMC - PubMed
-
- Guo Y, Sirkis DW, Schekman R. Protein Sorting at the trans-Golgi network. Annu Rev Cell Dev Biol. 2014;30:169–206. Epub 2014/08/26. doi: 10.1146/annurev-cellbio-100913-013012 - DOI - PubMed
-
- Babst M. Quality control: quality control at the plasma membrane: one mechanism does not fit all. J Cell Biol. 2014;205(1):11–20. Epub 2014/04/16. PubMed Central PMCID: PMCPmc3987138. doi: 10.1083/jcb.201310113 - DOI - PMC - PubMed
-
- Koenig PA, Ploegh HL. Protein quality control in the endoplasmic reticulum. F1000Prime Rep. 2014;6:49 Epub 2014/09/04. PubMed Central PMCID: PMCPmc4108957. doi: 10.12703/P6-49 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

