Genome wide profiling in oral squamous cell carcinoma identifies a four genetic marker signature of prognostic significance
- PMID: 28384287
- PMCID: PMC5383235
- DOI: 10.1371/journal.pone.0174865
Genome wide profiling in oral squamous cell carcinoma identifies a four genetic marker signature of prognostic significance
Abstract
Background: Cancers of the oral cavity are primarily oral squamous cell carcinomas (OSCCs). Many of the OSCCs present at late stages with an exceptionally poor prognosis. A probable limitation in management of patients with OSCC lies in the insufficient knowledge pertaining to the linkage between copy number alterations in OSCC and oral tumourigenesis thereby resulting in an inability to deliver targeted therapy.
Objectives: The current study aimed to identify copy number alterations (CNAs) in OSCC using array comparative genomic hybridization (array CGH) and to correlate the CNAs with clinico-pathologic parameters and clinical outcomes.
Materials and methods: Using array CGH, genome-wide profiling was performed on 75 OSCCs. Selected genes that were harboured in the frequently amplified and deleted regions were validated using quantitative polymerase chain reaction (qPCR). Thereafter, pathway and network functional analysis were carried out using Ingenuity Pathway Analysis (IPA) software.
Results: Multiple chromosomal regions including 3q, 5p, 7p, 8q, 9p, 10p, 11q were frequently amplified, while 3p and 8p chromosomal regions were frequently deleted. These findings were in confirmation with our previous study using ultra-dense array CGH. In addition, amplification of 8q, 11q, 7p and 9p and deletion of 8p chromosomal regions showed a significant correlation with clinico-pathologic parameters such as the size of the tumour, metastatic lymph nodes and pathological staging. Co-amplification of 7p, 8q, 9p and 11q regions that harbored amplified genes namely CCND1, EGFR, TPM2 and LRP12 respectively, when combined, continues to be an independent prognostic factor in OSCC.
Conclusion: Amplification of 3q, 5p, 7p, 8q, 9p, 10p, 11q and deletion of 3p and 8p chromosomal regions were recurrent among OSCC patients. Co-alteration of 7p, 8q, 9p and 11q was found to be associated with clinico-pathologic parameters and poor survival. These regions contain genes that play critical roles in tumourigenesis pathways.
Conflict of interest statement
Figures


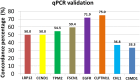

Similar articles
-
Genome-wide analysis of oral squamous cell carcinomas revealed over expression of ISG15, Nestin and WNT11.Oral Dis. 2012 Jul;18(5):469-76. doi: 10.1111/j.1601-0825.2011.01894.x. Epub 2012 Jan 18. Oral Dis. 2012. PMID: 22251088
-
Genomic DNA copy number alterations from precursor oral lesions to oral squamous cell carcinoma.Oral Oncol. 2014 May;50(5):404-12. doi: 10.1016/j.oraloncology.2014.02.005. Epub 2014 Mar 7. Oral Oncol. 2014. PMID: 24613650 Review.
-
Genetic differences detected by comparative genomic hybridization in head and neck squamous cell carcinomas from different tumor sites: construction of oncogenetic trees for tumor progression.Genes Chromosomes Cancer. 2002 Jun;34(2):224-33. doi: 10.1002/gcc.10062. Genes Chromosomes Cancer. 2002. PMID: 11979556
-
Genome-wide profiling of oral squamous cell carcinoma by array-based comparative genomic hybridization.Laryngoscope. 2006 May;116(5):735-41. doi: 10.1097/01.mlg.0000205141.54471.7f. Laryngoscope. 2006. PMID: 16652080
-
Chromosomal imbalances in oral squamous cell carcinoma: examination of 31 cell lines and review of the literature.Oral Oncol. 2008 Apr;44(4):369-82. doi: 10.1016/j.oraloncology.2007.05.003. Epub 2007 Aug 2. Oral Oncol. 2008. PMID: 17681875 Free PMC article. Review.
Cited by
-
Prediction of survival of HPV16-negative, p16-negative oral cavity cancer patients using a 13-gene signature: A multicenter study using FFPE samples.Oral Oncol. 2020 Jan;100:104487. doi: 10.1016/j.oraloncology.2019.104487. Epub 2019 Dec 10. Oral Oncol. 2020. PMID: 31835136 Free PMC article.
-
A Panel of Genes Identified as Targets for 8q24.13-24.3 Gain Contributing to Unfavorable Overall Survival in Patients with Hepatocellular Carcinoma.Curr Med Sci. 2018 Aug;38(4):590-596. doi: 10.1007/s11596-018-1918-x. Epub 2018 Aug 20. Curr Med Sci. 2018. PMID: 30128866
-
LTBP3 promotes early metastatic events during cancer cell dissemination.Oncogene. 2018 Apr;37(14):1815-1829. doi: 10.1038/s41388-017-0075-1. Epub 2018 Jan 19. Oncogene. 2018. PMID: 29348457 Free PMC article.
-
Tropomyosin 2 exerts anti-tumor effects in lung adenocarcinoma and is a novel prognostic biomarker.Histol Histopathol. 2023 Jun;38(6):669-680. doi: 10.14670/HH-18-514. Epub 2022 Jun 14. Histol Histopathol. 2023. PMID: 36102257
-
Patient-derived xenografts: a promising resource for preclinical cancer research.Mol Cell Oncol. 2019 Jan 8;6(1):1558684. doi: 10.1080/23723556.2018.1558684. eCollection 2019. Mol Cell Oncol. 2019. PMID: 30788424 Free PMC article.
References
-
- Ariffin OZ, Saleha IN. NCR report 2007. Ministry of Health, Malaysia. 2011.
-
- Silverman S Jr. Demographics and occurrence of oral and pharyngeal cancers. The outcomes, the trends, the challenge. J Am Dent Assoc. 2001;132 Suppl:7S–11S. Epub 2002/01/24. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

