Ziploc-ing the structure 2.0: Endoplasmic reticulum-resident peptidyl prolyl isomerases show different activities toward hydroxyproline
- PMID: 28385890
- PMCID: PMC5454108
- DOI: 10.1074/jbc.M116.772657
Ziploc-ing the structure 2.0: Endoplasmic reticulum-resident peptidyl prolyl isomerases show different activities toward hydroxyproline
Abstract
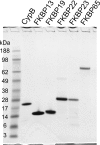
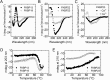
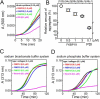
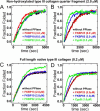
Extracellular matrix proteins are biosynthesized in the rough endoplasmic reticulum (rER), and the triple-helical protein collagen is the most abundant extracellular matrix component in the human body. Many enzymes, molecular chaperones, and post-translational modifiers facilitate collagen biosynthesis. Collagen contains a large number of proline residues, so the cis/trans isomerization of proline peptide bonds is the rate-limiting step during triple-helix formation. Accordingly, the rER-resident peptidyl prolyl cis/trans isomerases (PPIases) play an important role in the zipper-like triple-helix formation in collagen. We previously described this process as "Ziploc-ing the structure" and now provide additional information on the activity of individual rER PPIases. We investigated the substrate preferences of these PPIases in vitro using type III collagen, the unhydroxylated quarter fragment of type III collagen, and synthetic peptides as substrates. We observed changes in activity of six rER-resident PPIases, cyclophilin B (encoded by the PPIB gene), FKBP13 (FKBP2), FKBP19 (FKBP11), FKBP22 (FKBP14), FKBP23 (FKBP7), and FKBP65 (FKBP10), due to posttranslational modifications of proline residues in the substrate. Cyclophilin B and FKBP13 exhibited much lower activity toward post-translationally modified substrates. In contrast, FKBP19, FKBP22, and FKBP65 showed increased activity toward hydroxyproline-containing peptide substrates. Moreover, FKBP22 showed a hydroxyproline-dependent effect by increasing the amount of refolded type III collagen in vitro and FKBP19 seems to interact with triple helical type I collagen. Therefore, we propose that hydroxyproline modulates the rate of Ziploc-ing of the triple helix of collagen in the rER.
Keywords: biosynthesis; collagen; endoplasmic reticulum (ER); molecular chaperone; post-translational modification (PM); prolyl isomerase.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no competing interests related to this work
Figures
Similar articles
-
A substrate preference for the rough endoplasmic reticulum resident protein FKBP22 during collagen biosynthesis.J Biol Chem. 2014 Jun 27;289(26):18189-201. doi: 10.1074/jbc.M114.561944. Epub 2014 May 12. J Biol Chem. 2014. PMID: 24821723 Free PMC article.
-
Ziploc-ing the structure: Triple helix formation is coordinated by rough endoplasmic reticulum resident PPIases.Biochim Biophys Acta. 2015 Oct;1850(10):1983-93. doi: 10.1016/j.bbagen.2014.12.024. Epub 2015 Jan 10. Biochim Biophys Acta. 2015. PMID: 25583561 Review.
-
Structure of human peptidyl-prolyl cis-trans isomerase FKBP22 containing two EF-hand motifs.Protein Sci. 2014 Jan;23(1):67-75. doi: 10.1002/pro.2391. Epub 2013 Nov 25. Protein Sci. 2014. PMID: 24272907 Free PMC article.
-
Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes.Hum Mol Genet. 2011 Apr 15;20(8):1595-609. doi: 10.1093/hmg/ddr037. Epub 2011 Jan 31. Hum Mol Genet. 2011. PMID: 21282188 Free PMC article.
-
FK506-Binding protein 22 from a psychrophilic bacterium, a cold shock-inducible peptidyl prolyl isomerase with the ability to assist in protein folding.Int J Mol Sci. 2011;12(8):5261-84. doi: 10.3390/ijms12085261. Epub 2011 Aug 17. Int J Mol Sci. 2011. PMID: 21954357 Free PMC article. Review.
Cited by
-
Is Exon Skipping a Viable Therapeutic Approach for Vascular Ehlers-Danlos Syndrome with Mutations in COL3A1 Exon 10 or 15?Int J Mol Sci. 2024 Aug 13;25(16):8816. doi: 10.3390/ijms25168816. Int J Mol Sci. 2024. PMID: 39201504 Free PMC article.
-
FK506-Binding Protein 13 Expression Is Upregulated in Interstitial Lung Disease and Correlated with Clinical Severity. A Potentially Protective Role.Am J Respir Cell Mol Biol. 2021 Feb;64(2):235-246. doi: 10.1165/rcmb.2020-0121OC. Am J Respir Cell Mol Biol. 2021. PMID: 33253593 Free PMC article.
-
Promoting collagen synthesis: a viable strategy to combat skin ageing.J Enzyme Inhib Med Chem. 2025 Dec;40(1):2488821. doi: 10.1080/14756366.2025.2488821. Epub 2025 Apr 11. J Enzyme Inhib Med Chem. 2025. PMID: 40213810 Free PMC article. Review.
-
The Ehlers-Danlos Syndromes against the Backdrop of Inborn Errors of Metabolism.Genes (Basel). 2022 Jan 29;13(2):265. doi: 10.3390/genes13020265. Genes (Basel). 2022. PMID: 35205310 Free PMC article. Review.
-
Compartmentalized Proteomic Profiling Outlines the Crucial Role of the Classical Secretory Pathway during Recombinant Protein Production in Chinese Hamster Ovary Cells.ACS Omega. 2021 May 3;6(19):12439-12458. doi: 10.1021/acsomega.0c06030. eCollection 2021 May 18. ACS Omega. 2021. PMID: 34056395 Free PMC article.
References
-
- Szabadkai G., and Rizzuto R. (2007) Chaperones as parts of organelle networks. Adv. Exp. Med. Biol. 594, 64–77 - PubMed
-
- Hartl F. U., Bracher A., and Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 - PubMed
-
- Braakman I., and Bulleid N. J. (2011) Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 80, 71–99 - PubMed
-
- Bächinger H. P., Mizuno K., Vranka J. A., and Boudko S. P. (2010) Collagen formation and structure. In Comprehensive Natural Products II-Chemistry and Biology (Mander L., and Lui H. W., eds) pp 469–530, Elsevier, Oxford
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

