Elucidating dynamic metabolic physiology through network integration of quantitative time-course metabolomics
- PMID: 28387366
- PMCID: PMC5384226
- DOI: 10.1038/srep46249
Elucidating dynamic metabolic physiology through network integration of quantitative time-course metabolomics
Abstract
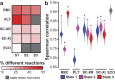
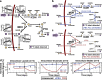
The increasing availability of metabolomics data necessitates novel methods for deeper data analysis and interpretation. We present a flux balance analysis method that allows for the computation of dynamic intracellular metabolic changes at the cellular scale through integration of time-course absolute quantitative metabolomics. This approach, termed "unsteady-state flux balance analysis" (uFBA), is applied to four cellular systems: three dynamic and one steady-state as a negative control. uFBA and FBA predictions are contrasted, and uFBA is found to be more accurate in predicting dynamic metabolic flux states for red blood cells, platelets, and Saccharomyces cerevisiae. Notably, only uFBA predicts that stored red blood cells metabolize TCA intermediates to regenerate important cofactors, such as ATP, NADH, and NADPH. These pathway usage predictions were subsequently validated through 13C isotopic labeling and metabolic flux analysis in stored red blood cells. Utilizing time-course metabolomics data, uFBA provides an accurate method to predict metabolic physiology at the cellular scale for dynamic systems.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

