Proximity-dependent labeling methods for proteomic profiling in living cells
- PMID: 28387482
- PMCID: PMC5553119
- DOI: 10.1002/wdev.272
Proximity-dependent labeling methods for proteomic profiling in living cells
Abstract
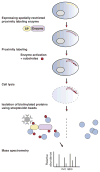
Characterizing the proteome composition of organelles and subcellular regions of living cells can facilitate the understanding of cellular organization as well as protein interactome networks. Proximity labeling-based methods coupled with mass spectrometry (MS) offer a high-throughput approach for systematic analysis of spatially restricted proteomes. Proximity labeling utilizes enzymes that generate reactive radicals to covalently tag neighboring proteins with biotin. The biotinylated endogenous proteins can then be isolated for further analysis by MS. To analyze protein-protein interactions or identify components that localize to discrete subcellular compartments, spatial expression is achieved by fusing the enzyme to specific proteins or signal peptides that target to particular subcellular regions. Although these technologies have only been introduced recently, they have already provided deep insights into a wide range of biological processes. Here, we describe and compare current methods of proximity labeling as well as their applications. As each method has its own unique features, the goal of this review is to describe how different proximity labeling methods can be used to answer different biological questions. WIREs Dev Biol 2017, 6:e272. doi: 10.1002/wdev.272 For further resources related to this article, please visit the WIREs website.
© 2017 Wiley Periodicals, Inc.
Conflict of interest statement
Conflict of interest: The authors have declared no conflicts of interest for this article.
Figures

References
-
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

