Phenome-wide heritability analysis of the UK Biobank
- PMID: 28388634
- PMCID: PMC5400281
- DOI: 10.1371/journal.pgen.1006711
Phenome-wide heritability analysis of the UK Biobank
Erratum in
-
Correction: Phenome-wide heritability analysis of the UK Biobank.PLoS Genet. 2018 Feb 9;14(2):e1007228. doi: 10.1371/journal.pgen.1007228. eCollection 2018 Feb. PLoS Genet. 2018. PMID: 29425192 Free PMC article.
Abstract
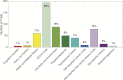
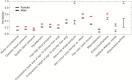
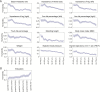
Heritability estimation provides important information about the relative contribution of genetic and environmental factors to phenotypic variation, and provides an upper bound for the utility of genetic risk prediction models. Recent technological and statistical advances have enabled the estimation of additive heritability attributable to common genetic variants (SNP heritability) across a broad phenotypic spectrum. Here, we present a computationally and memory efficient heritability estimation method that can handle large sample sizes, and report the SNP heritability for 551 complex traits derived from the interim data release (152,736 subjects) of the large-scale, population-based UK Biobank, comprising both quantitative phenotypes and disease codes. We demonstrate that common genetic variation contributes to a broad array of quantitative traits and human diseases in the UK population, and identify phenotypes whose heritability is moderated by age (e.g., a majority of physical measures including height and body mass index), sex (e.g., blood pressure related traits) and socioeconomic status (education). Our study represents the first comprehensive phenome-wide heritability analysis in the UK Biobank, and underscores the importance of considering population characteristics in interpreting heritability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Visscher P. M., Hill W. G., Wray N. R. (2008). Heritability in the genomics era—concepts and misconceptions. Nature Reviews Genetics, 9(4), 255–266. doi: 10.1038/nrg2322 - DOI - PubMed
-
- Polderman T. J., Benyamin B., De Leeuw C. A., Sullivan P. F., Van Bochoven A., Visscher P. M., et al. (2015). Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nature Genetics, 47(7), 702–709. doi: 10.1038/ng.3285 - DOI - PubMed
-
- Lee S. H., Wray N. R., Goddard M. E., Visscher P. M. (2011). Estimating missing heritability for disease from genome-wide association studies. The American Journal of Human Genetics, 88(3), 294–305. doi: 10.1016/j.ajhg.2011.02.002 - DOI - PMC - PubMed
-
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., Nyholt D. R., et al. (2010). Common SNPs explain a large proportion of the heritability for human height. Nature Genetics, 42(7), 565–569. doi: 10.1038/ng.608 - DOI - PMC - PubMed
-
- Yang J., Lee S. H., Goddard M. E., Visscher P. M. (2011). GCTA: a tool for genome-wide complex trait analysis. The American Journal of Human Genetics, 88(1), 76–82. doi: 10.1016/j.ajhg.2010.11.011 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

