Characterization and mutational analysis of a nicotinamide mononucleotide deamidase from Agrobacterium tumefaciens showing high thermal stability and catalytic efficiency
- PMID: 28388636
- PMCID: PMC5384747
- DOI: 10.1371/journal.pone.0174759
Characterization and mutational analysis of a nicotinamide mononucleotide deamidase from Agrobacterium tumefaciens showing high thermal stability and catalytic efficiency
Abstract
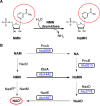
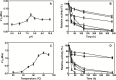
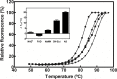
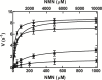
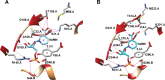
NAD+ has emerged as a crucial element in both bioenergetic and signaling pathways since it acts as a key regulator of cellular and organismal homeostasis. Among the enzymes involved in its recycling, nicotinamide mononucleotide (NMN) deamidase is one of the key players in the bacterial pyridine nucleotide cycle, where it catalyzes the conversion of NMN into nicotinic acid mononucleotide (NaMN), which is later converted to NAD+ in the Preiss-Handler pathway. The biochemical characteristics of bacterial NMN deamidases have been poorly studied, although they have been investigated in some firmicutes, gamma-proteobacteria and actinobacteria. In this study, we present the first characterization of an NMN deamidase from an alphaproteobacterium, Agrobacterium tumefaciens (AtCinA). The enzyme was active over a broad pH range, with an optimum at pH 7.5. Moreover, the enzyme was quite stable at neutral pH, maintaining 55% of its activity after 14 days. Surprisingly, AtCinA showed the highest optimal (80°C) and melting (85°C) temperatures described for an NMN deamidase. The above described characteristics, together with its high catalytic efficiency, make AtCinA a promising biocatalyst for the production of pure NaMN. In addition, six mutants (C32A, S48A, Y58F, Y58A, T105A and R145A) were designed to study their involvement in substrate binding, and two (S31A and K63A) to determine their contribution to the catalysis. However, only four mutants (C32A, S48A Y58F and T105A) showed activity, although with reduced catalytic efficiency. These results, combined with a thermal and structural analysis, reinforce the Ser/Lys catalytic dyad mechanism as the most plausible among those proposed.
Conflict of interest statement
Figures

Similar articles
-
Characterization of Two NMN Deamidase Mutants as Possible Probes for an NMN Biosensor.Int J Mol Sci. 2021 Jun 13;22(12):6334. doi: 10.3390/ijms22126334. Int J Mol Sci. 2021. PMID: 34199271 Free PMC article.
-
New insights into the phylogeny and molecular classification of nicotinamide mononucleotide deamidases.PLoS One. 2013 Dec 5;8(12):e82705. doi: 10.1371/journal.pone.0082705. eCollection 2013. PLoS One. 2013. PMID: 24340054 Free PMC article.
-
Characterization of bacterial NMN deamidase as a Ser/Lys hydrolase expands diversity of serine amidohydrolases.FEBS Lett. 2014 Mar 18;588(6):1016-23. doi: 10.1016/j.febslet.2014.01.063. Epub 2014 Feb 11. FEBS Lett. 2014. PMID: 24530526
-
Isolation of NAD cycle mutants defective in nicotinamide mononucleotide deamidase in Salmonella typhimurium.J Bacteriol. 1995 Dec;177(23):6711-7. doi: 10.1128/jb.177.23.6711-6717.1995. J Bacteriol. 1995. PMID: 7592458 Free PMC article.
-
Identification of nicotinamide mononucleotide deamidase of the bacterial pyridine nucleotide cycle reveals a novel broadly conserved amidohydrolase family.J Biol Chem. 2011 Nov 18;286(46):40365-75. doi: 10.1074/jbc.M111.275818. Epub 2011 Sep 27. J Biol Chem. 2011. PMID: 21953451 Free PMC article.
Cited by
-
Characterization of Two NMN Deamidase Mutants as Possible Probes for an NMN Biosensor.Int J Mol Sci. 2021 Jun 13;22(12):6334. doi: 10.3390/ijms22126334. Int J Mol Sci. 2021. PMID: 34199271 Free PMC article.
-
Comparative inhibitory profile and distribution of bacterial PARPs, using Clostridioides difficile CD160 PARP as a model.Sci Rep. 2018 May 23;8(1):8056. doi: 10.1038/s41598-018-26450-0. Sci Rep. 2018. PMID: 29795234 Free PMC article.
-
Characterization of Resveratrol, Oxyresveratrol, Piceatannol and Roflumilast as Modulators of Phosphodiesterase Activity. Study of Yeast Lifespan.Pharmaceuticals (Basel). 2020 Aug 30;13(9):225. doi: 10.3390/ph13090225. Pharmaceuticals (Basel). 2020. PMID: 32872677 Free PMC article.
-
An uncharacterized FMAG_01619 protein from Fusobacterium mortiferum ATCC 9817 demonstrates that some bacterial macrodomains can also act as poly-ADP-ribosylhydrolases.Sci Rep. 2019 Mar 1;9(1):3230. doi: 10.1038/s41598-019-39691-4. Sci Rep. 2019. PMID: 30824723 Free PMC article.
-
CinA mediates multidrug tolerance in Mycobacterium tuberculosis.Nat Commun. 2022 Apr 22;13(1):2203. doi: 10.1038/s41467-022-29832-1. Nat Commun. 2022. PMID: 35459278 Free PMC article.
References
-
- Butepage M, Eckei L, Verheugd P, Luscher B (2015) Intracellular Mono-ADP-Ribosylation in Signaling and Disease. Cells 4: 569–595. PubMed Central PMCID: PMC4695847. doi: 10.3390/cells4040569 - DOI - PMC - PubMed
-
- Verheugd P, Butepage M, Eckei L, Luscher B (2016) Players in ADP-ribosylation: readers and erasers. Curr Protein Pept Sci 17: 654–667. - PubMed
-
- Kupis W, Palyga J, Tomal E, Niewiadomska E (2016) The role of sirtuins in cellular homeostasis. J Physiol Biochem 72: 371–380. PubMed Central PMCID: PMC4992043. doi: 10.1007/s13105-016-0492-6 - DOI - PMC - PubMed
-
- Sorci L, Ruggieri S, Raffaelli N (2014) NAD homeostasis in the bacterial response to DNA/RNA damage. DNA Repair (Amst) 23: 17–26. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

