Combining clinical and genomics queries using i2b2 - Three methods
- PMID: 28388645
- PMCID: PMC5384666
- DOI: 10.1371/journal.pone.0172187
Combining clinical and genomics queries using i2b2 - Three methods
Abstract
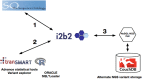
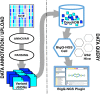
We are fortunate to be living in an era of twin biomedical data surges: a burgeoning representation of human phenotypes in the medical records of our healthcare systems, and high-throughput sequencing making rapid technological advances. The difficulty representing genomic data and its annotations has almost by itself led to the recognition of a biomedical "Big Data" challenge, and the complexity of healthcare data only compounds the problem to the point that coherent representation of both systems on the same platform seems insuperably difficult. We investigated the capability for complex, integrative genomic and clinical queries to be supported in the Informatics for Integrating Biology and the Bedside (i2b2) translational software package. Three different data integration approaches were developed: The first is based on Sequence Ontology, the second is based on the tranSMART engine, and the third on CouchDB. These novel methods for representing and querying complex genomic and clinical data on the i2b2 platform are available today for advancing precision medicine.
Conflict of interest statement
Figures

Similar articles
-
BigQ: a NoSQL based framework to handle genomic variants in i2b2.BMC Bioinformatics. 2015 Dec 29;16:415. doi: 10.1186/s12859-015-0861-0. BMC Bioinformatics. 2015. PMID: 26714792 Free PMC article.
-
Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside.AMIA Annu Symp Proc. 2007 Oct 11;2007:548-52. AMIA Annu Symp Proc. 2007. PMID: 18693896 Free PMC article.
-
Computing health quality measures using Informatics for Integrating Biology and the Bedside.J Med Internet Res. 2013 Apr 19;15(4):e75. doi: 10.2196/jmir.2493. J Med Internet Res. 2013. PMID: 23603227 Free PMC article.
-
Automating Installation of the Integrating Biology and the Bedside (i2b2) Platform.Biomed Inform Insights. 2018 Jun 4;10:1178222618777749. doi: 10.1177/1178222618777749. eCollection 2018. Biomed Inform Insights. 2018. PMID: 29887730 Free PMC article. Review.
-
From genome sequencing to bedside. Findings from the section on bioinformatics and translational informatics.Yearb Med Inform. 2013;8:175-7. Yearb Med Inform. 2013. PMID: 23974568 Review.
Cited by
-
Enabling Precision Medicine in Cancer Care Through a Molecular Data Warehouse: The Moffitt Experience.JCO Clin Cancer Inform. 2021 May;5:561-569. doi: 10.1200/CCI.20.00175. JCO Clin Cancer Inform. 2021. PMID: 33989014 Free PMC article.
-
The Association of Black Cardiologists (ABC) Cardiovascular Implementation Study (CVIS): A Research Registry Integrating Social Determinants to Support Care for Underserved Patients.Int J Environ Res Public Health. 2019 May 10;16(9):1631. doi: 10.3390/ijerph16091631. Int J Environ Res Public Health. 2019. PMID: 31083298 Free PMC article.
-
Research data warehouse best practices: catalyzing national data sharing through informatics innovation.J Am Med Inform Assoc. 2022 Mar 15;29(4):581-584. doi: 10.1093/jamia/ocac024. J Am Med Inform Assoc. 2022. PMID: 35289371 Free PMC article. No abstract available.
-
Phenotyping to Facilitate Accrual for a Cardiovascular Intervention.J Clin Med Res. 2019 Jun;11(6):458-463. doi: 10.14740/jocmr3830. Epub 2019 May 10. J Clin Med Res. 2019. PMID: 31143314 Free PMC article.
-
Integrating Genomics and Clinical Data for Statistical Analysis by Using GEnome MINIng (GEMINI) and Fast Healthcare Interoperability Resources (FHIR): System Design and Implementation.J Med Internet Res. 2020 Oct 7;22(10):e19879. doi: 10.2196/19879. J Med Internet Res. 2020. PMID: 33026356 Free PMC article.
References
-
- Kurreeman F.,Liao K., Chibnik L., Hickey B., Stahl E., Gainer V., et al., Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet, 2011. 88(1): p. 57–69. 10.1016/j.ajhg.2010.12.007 - DOI - PMC - PubMed
-
- Savaiano J. Bring healthcare's dark data to light. In: healthcareitnews.com [Internet]. 30 Jan 2013 [cited 20 Nov 2014]. http://www.healthcareitnews.com/news/bring-healthcares-dark-data-light?s...
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

