Dose-response relationships for environmentally mediated infectious disease transmission models
- PMID: 28388665
- PMCID: PMC5400279
- DOI: 10.1371/journal.pcbi.1005481
Dose-response relationships for environmentally mediated infectious disease transmission models
Erratum in
-
Correction: Dose-response relationships for environmentally mediated infectious disease transmission models.PLoS Comput Biol. 2017 Sep 20;13(9):e1005765. doi: 10.1371/journal.pcbi.1005765. eCollection 2017 Sep. PLoS Comput Biol. 2017. PMID: 28931005 Free PMC article.
Abstract
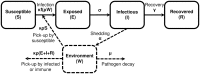
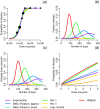
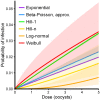
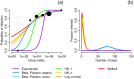
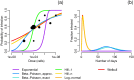
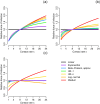
Environmentally mediated infectious disease transmission models provide a mechanistic approach to examining environmental interventions for outbreaks, such as water treatment or surface decontamination. The shift from the classical SIR framework to one incorporating the environment requires codifying the relationship between exposure to environmental pathogens and infection, i.e. the dose-response relationship. Much of the work characterizing the functional forms of dose-response relationships has used statistical fit to experimental data. However, there has been little research examining the consequences of the choice of functional form in the context of transmission dynamics. To this end, we identify four properties of dose-response functions that should be considered when selecting a functional form: low-dose linearity, scalability, concavity, and whether it is a single-hit model. We find that i) middle- and high-dose data do not constrain the low-dose response, and different dose-response forms that are equally plausible given the data can lead to significant differences in simulated outbreak dynamics; ii) the choice of how to aggregate continuous exposure into discrete doses can impact the modeled force of infection; iii) low-dose linear, concave functions allow the basic reproduction number to control global dynamics; and iv) identifiability analysis offers a way to manage multiple sources of uncertainty and leverage environmental monitoring to make inference about infectivity. By applying an environmentally mediated infectious disease model to the 1993 Milwaukee Cryptosporidium outbreak, we demonstrate that environmental monitoring allows for inference regarding the infectivity of the pathogen and thus improves our ability to identify outbreak characteristics such as pathogen strain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Capasso V, Paveri-Fontana SL. A mathematical model for the 1973 cholera epidemic in the European Mediterranean region. Revue d’épidémiologie et de santé publique. 1979;27(2):121–32. - PubMed
-
- Bertuzzo E, Azaele S, Maritan A, Gatto M, Rodriguez-Iturbe I, Rinaldo A. On the space-time evolution of a cholera epidemic. Water Resources Research. 2008;44(1):1–8. 10.1029/2007WR006211 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

