Experimental Evaluation of Host Adaptation of Lactobacillus reuteri to Different Vertebrate Species
- PMID: 28389535
- PMCID: PMC5452824
- DOI: 10.1128/AEM.00132-17
Experimental Evaluation of Host Adaptation of Lactobacillus reuteri to Different Vertebrate Species
Abstract
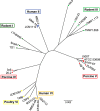
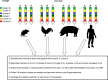
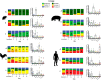
The species Lactobacillus reuteri has diversified into host-specific lineages, implying a long-term association with different vertebrates. Strains from rodent lineages show specific adaptations to mice, but the processes underlying the evolution of L. reuteri in other hosts remain unknown. We administered three standardized inocula composed of strains from different host-confined lineages to mice, pigs, chickens, and humans. The ecological performance of each strain in the gastrointestinal tract of each host was determined by typing random colonies recovered from fecal samples collected over five consecutive days postadministration. Results revealed that rodent strains were predominant in mice, confirming previous findings of host adaptation. In chickens, poultry strains of the lineage VI (poultry VI) and human isolates from the same lineage (human VI) were recovered at the highest and second highest rates, respectively. Interestingly, human VI strains were virtually undetected in human feces. These findings, together with ancestral state reconstructions, indicate poultry VI and human VI strains share an evolutionary history with chickens. Genomic analysis revealed that poultry VI strains possess a large and variable accessory genome, whereas human VI strains display low genetic diversity and possess genes encoding antibiotic resistance and capsular polysaccharide synthesis, which might have allowed temporal colonization of humans. Experiments in pigs and humans did not provide evidence of host adaptation of L. reuteri to these hosts. Overall, our findings demonstrate host adaptation of L. reuteri to rodents and chickens, supporting a joint evolution of this bacterial species with several vertebrate hosts, although questions remain about its natural history in humans and pigs.IMPORTANCE Gut microbes are often hypothesized to have coevolved with their vertebrate hosts. However, the evidence is sparse and the evolutionary mechanisms have not been identified. We developed and applied an experimental approach to determine host adaptation of L. reuteri to different hosts. Our findings confirmed adaptation to rodents and provided evidence of adaptation to poultry, suggesting that L. reuteri evolved via natural selection in different hosts. By complementing phylogenetic analyses with experimental evidence, this study provides novel information about the mechanisms driving host-microbe coevolution with vertebrates and serve as a basis to inform the application of L. reuteri as a probiotic for different host species.
Keywords: Lactobacillus reuteri; host adaptation; probiotics; symbiosis.
Copyright © 2017 American Society for Microbiology.
Figures
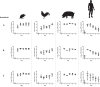

References
-
- Stevens CE, Huma ID. 1998. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol Rev 78:393–427. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

