Microbiota in Exhaled Breath Condensate and the Lung
- PMID: 28389539
- PMCID: PMC5452816
- DOI: 10.1128/AEM.00515-17
Microbiota in Exhaled Breath Condensate and the Lung
Abstract
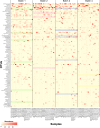
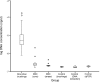
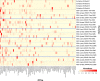
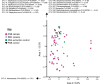
The lung microbiota is commonly sampled using relatively invasive bronchoscopic procedures. Exhaled breath condensate (EBC) collection potentially offers a less invasive alternative for lung microbiota sampling. We compared lung microbiota samples retrieved by protected specimen brushings (PSB) and exhaled breath condensate collection. We also sought to assess whether aerosolized antibiotic treatment would influence the lung microbiota and whether this change could be detected in EBC. EBC was collected from 6 conscious sheep and then from the same anesthetized sheep during mechanical ventilation. Following the latter EBC collection, PSB samples were collected from separate sites within each sheep lung. On the subsequent day, each sheep was then treated with nebulized colistimethate sodium. Two days after nebulization, EBC and PSB samples were again collected. Bacterial DNA was quantified using 16S rRNA gene quantitative PCR. The V2-V3 region of the 16S rRNA gene was amplified by PCR and sequenced using Illumina MiSeq. Quality control and operational taxonomic unit (OTU) clustering were performed with mothur. The EBC samples contained significantly less bacterial DNA than the PSB samples. The EBC samples from anesthetized animals clustered separately by their bacterial community compositions in comparison to the PSB samples, and 37 bacterial OTUs were identified as differentially abundant between the two sample types. Despite only low concentrations of colistin being detected in bronchoalveolar lavage fluid, PSB samples were found to differ by their bacterial compositions before and after colistimethate sodium treatment. Our findings indicate that microbiota in EBC samples and PSB samples are not equivalent.IMPORTANCE Sampling of the lung microbiota usually necessitates performing bronchoscopic procedures that involve a hospital visit for human participants and the use of trained staff. The inconvenience and perceived discomfort of participating in this kind of research may deter healthy volunteers and may not be a safe option for patients with advanced lung disease. This study set out to evaluate a less invasive method for collecting lung microbiota samples by comparing samples taken via protected specimen brushings (PSB) to those taken via exhaled breath condensate (EBC) collection. We found that there was less bacterial DNA in EBC samples compared with that in PSB samples and that there were differences between the bacterial communities in the two sample types. We conclude that while EBC and PSB samples do not produce equivalent microbiota samples, the study of the EBC microbiota may still be of interest.
Keywords: 16S rRNA; EBC; colistimethate sodium; colistin; lung microbiota; respiratory microbiota; sheep.
Copyright © 2017 Glendinning et al.
Figures
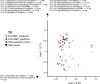
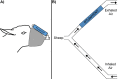

References
-
- Bhimji A, Singer LG, Kumar D, Humar A, Pavan R, Zhang H, Rotstein C, Keshavjee S, Mazzulli T, Husain S. 2016. Feasibility of detecting fungal DNA in exhaled breath condensate by the Luminex Multiplex xTAG fungal PCR assay in lung transplant recipients: a pilot study. J Heart Lung Transplant 35:S37. doi:10.1016/j.healun.2016.01.099. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

