Acid Evolution of Escherichia coli K-12 Eliminates Amino Acid Decarboxylases and Reregulates Catabolism
- PMID: 28389540
- PMCID: PMC5452808
- DOI: 10.1128/AEM.00442-17
Acid Evolution of Escherichia coli K-12 Eliminates Amino Acid Decarboxylases and Reregulates Catabolism
Abstract
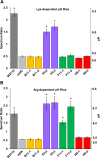
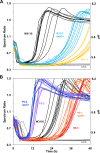
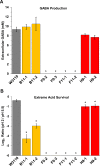
Acid-adapted strains of Escherichia coli K-12 W3110 were obtained by serial culture in medium buffered at pH 4.6 (M. M. Harden, A. He, K. Creamer, M. W. Clark, I. Hamdallah, K. A. Martinez, R. L. Kresslein, S. P. Bush, and J. L. Slonczewski, Appl Environ Microbiol 81:1932-1941, 2015, https://doi.org/10.1128/AEM.03494-14). Revised genomic analysis of these strains revealed insertion sequence (IS)-driven insertions and deletions that knocked out regulators CadC (acid induction of lysine decarboxylase), GadX (acid induction of glutamate decarboxylase), and FNR (anaerobic regulator). Each acid-evolved strain showed loss of one or more amino acid decarboxylase systems, which normally help neutralize external acid (pH 5 to 6) and increase survival in extreme acid (pH 2). Strains from populations B11, H9, and F11 had an IS5 insertion or IS-mediated deletion in cadC, while population B11 had a point mutation affecting the arginine activator adiY The cadC and adiY mutants failed to neutralize acid in the presence of exogenous lysine or arginine. In strain B11-1, reversion of an rpoC (RNA polymerase) mutation partly restored arginine-dependent neutralization. All eight strains showed deletion or downregulation of the Gad acid fitness island. Strains with the Gad deletion lost the ability to produce GABA (gamma-aminobutyric acid) and failed to survive extreme acid. Transcriptome sequencing (RNA-seq) of strain B11-1 showed upregulated genes for catabolism of diverse substrates but downregulated acid stress genes (the biofilm regulator ariR, yhiM, and Gad). Other strains showed downregulation of H2 consumption mediated by hydrogenases (hya and hyb) which release acid. Strains F9-2 and F9-3 had a deletion of fnr and showed downregulation of FNR-dependent genes (dmsABC, frdABCD, hybABO, nikABCDE, and nrfAC). Overall, strains that had evolved in buffered acid showed loss or downregulation of systems that neutralize unbuffered acid and showed altered regulation of catabolism.IMPORTANCE Experimental evolution of an enteric bacterium under a narrow buffered range of acid pH leads to loss of genes that enhance fitness above or below the buffered pH range, including loss of enzymes that may raise external pH in the absence of buffer. Prominent modes of evolutionary change involve IS-mediated insertions and deletions that knock out key regulators. Over generations of acid stress, catabolism undergoes reregulation in ways that differ for each evolving strain.
Keywords: Escherichia coli; GABA; RNA polymerase; acid; decarboxylase; experimental evolution; fnr; low pH.
Copyright © 2017 American Society for Microbiology.
Figures
References
-
- Fallingborg J. 1999. Intraluminal pH of the human gastrointestinal tract. Dan Med Bull 46:183–196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

