Large-scale analysis of variation in the insulin-like growth factor family in humans reveals rare disease links and common polymorphisms
- PMID: 28389567
- PMCID: PMC5454106
- DOI: 10.1074/jbc.M117.783639
Large-scale analysis of variation in the insulin-like growth factor family in humans reveals rare disease links and common polymorphisms
Erratum in
-
Large-scale analysis of variation in the insulin-like growth factor family in humans reveals rare disease links and common polymorphisms.J Biol Chem. 2017 Dec 1;292(48):19608. doi: 10.1074/jbc.AAC117.000854. J Biol Chem. 2017. PMID: 29196571 Free PMC article. No abstract available.
Abstract
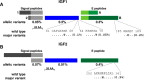
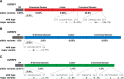
The insulin-like growth factors IGF1 and IGF2 are closely related proteins that are essential for normal growth and development in humans and other species and play critical roles in many physiological and pathophysiological processes. IGF actions are mediated by transmembrane receptors and modulated by IGF-binding proteins. The importance of IGF actions in human physiology is strengthened by the rarity of inactivating mutations in their genes and by the devastating impact caused by such mutations on normal development and somatic growth. Large-scale genome sequencing has the potential to provide new insights into human variation and disease susceptibility. Toward this end, the availability of DNA sequence data from 60,706 people through the Exome Aggregation Consortium has prompted the analyses presented here. Results reveal a broad range of potential missense and other alterations in the coding regions of every IGF family gene, but the vast majority of predicted changes were uncommon. The total number of different alleles detected per gene in the population varied over an ∼15-fold range, from 57 for IGF1 to 872 for IGF2R, although when corrected for protein length the rate ranged from 0.22 to 0.59 changes/codon among the 11 genes evaluated. Previously characterized disease-causing mutations in IGF2, IGF1R, IGF2R, or IGFALS all were found in the general population but with allele frequencies of <1:30,000. A few new highly prevalent amino acid polymorphisms were also identified. Collectively, these data provide a wealth of opportunities to understand the intricacies of IGF signaling and action in both physiological and pathological contexts.
Keywords: IGF binding proteins; IGF receptor; functional genomics; genomics; growth factor; human evolution; human variation; insulin; insulin-like growth factor (IGF); population genetics.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The author declares that he has no conflicts of interest with the contents of this article
Figures



References
-
- Le Roith D., Bondy C., Yakar S., Liu J. L., and Butler A. (2001) The somatomedin hypothesis: 2001. Endocr. Rev. 22, 53–74 - PubMed
-
- LeRoith D. (2008) Clinical relevance of systemic and local IGF-I: lessons from animal models. Pediatr. Endocrinol. Rev. 5, 739–743 - PubMed
-
- Pollak M. (2012) The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat. Rev. Cancer 12, 159–169 - PubMed
-
- Gems D., and Partridge L. (2013) Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol. 75, 621–644 - PubMed
-
- Stewart C. E., and Rotwein P. (1996) Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol. Rev. 76, 1005. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

