Cancer Immunogenomics: Computational Neoantigen Identification and Vaccine Design
- PMID: 28389595
- PMCID: PMC5702270
- DOI: 10.1101/sqb.2016.81.030726
Cancer Immunogenomics: Computational Neoantigen Identification and Vaccine Design
Abstract
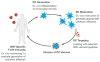
The application of modern high-throughput genomics to the study of cancer genomes has exploded in the past few years, yielding unanticipated insights into the myriad and complex combinations of genomic alterations that lead to the development of cancers. Coincident with these genomic approaches have been computational analyses that are capable of multiplex evaluations of genomic data toward specific therapeutic end points. One such approach is called "immunogenomics" and is now being developed to interpret protein-altering changes in cancer cells in the context of predicted preferential binding of these altered peptides by the patient's immune molecules, specifically human leukocyte antigen (HLA) class I and II proteins. One goal of immunogenomics is to identify those cancer-specific alterations that are likely to elicit an immune response that is highly specific to the patient's cancer cells following stimulation by a personalized vaccine. The elements of such an approach are outlined herein and constitute an emerging therapeutic option for cancer patients.
© 2016 Hundal et al; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. 1985. J Immunol. 2005;175:4163–4165. - PubMed
-
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
