RNA sequence analysis of rat acute experimental pancreatitis with and without fatty liver: a gene expression profiling comparative study
- PMID: 28389636
- PMCID: PMC5429720
- DOI: 10.1038/s41598-017-00821-5
RNA sequence analysis of rat acute experimental pancreatitis with and without fatty liver: a gene expression profiling comparative study
Abstract
Fatty liver (FL) is one of the risk factors for acute pancreatitis and is also indicative of a worse prognosis as compared to acute pancreatitis without fatty liver (AP). The aim of the present study was to analyze, at the hepatic level, the differentially expressed genes (DEGs) between acute pancreatitis with fatty liver (APFL) rats and AP rats. GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analyses of these DEGs indicated that PPARα signalling pathway and fatty acid degradation pathway may be involved in the pathological process of APFL, which indicated that fatty liver may aggravate pancreatitis through these pathways. Moreover, the excessive activation of JAK/STAT signaling pathway and toll-like receptor signaling pathway was also found in APFL group as shown in heat map. In conclusion, the inhibition of PPARα signaling pathway and the fatty acid degradation pathway may lead to the further disorder of lipid metabolism, which can aggravate pancreatitis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


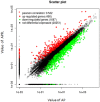




 ), JAK/STAT signaling pathway were marked with an asterisk (
), JAK/STAT signaling pathway were marked with an asterisk ( ), endoplasmic reticulum stress were marked with an asterisk (
), endoplasmic reticulum stress were marked with an asterisk ( ), chemokine receptors were marked with an asterisk (
), chemokine receptors were marked with an asterisk ( ), tumor necrosis factor receptor superfamily were marked with an asterisk (
), tumor necrosis factor receptor superfamily were marked with an asterisk ( ), interleukin were marked with an asterisk (
), interleukin were marked with an asterisk ( ) and toll-like receptors were marked with an asterisk (
) and toll-like receptors were marked with an asterisk ( ).
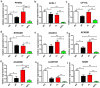
).
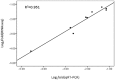

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

