Biochemical analysis of leishmanial and human GDP-Mannose Pyrophosphorylases and selection of inhibitors as new leads
- PMID: 28389670
- PMCID: PMC5429698
- DOI: 10.1038/s41598-017-00848-8
Biochemical analysis of leishmanial and human GDP-Mannose Pyrophosphorylases and selection of inhibitors as new leads
Abstract
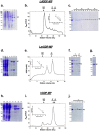
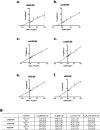
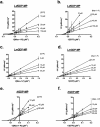
Leishmaniases are an ensemble of diseases caused by the protozoan parasite of the genus Leishmania. Current antileishmanial treatments are limited and present main issues of toxicity and drug resistance emergence. Therefore, the generation of new inhibitors specifically directed against a leishmanial target is an attractive strategy to expand the chemotherapeutic arsenal. GDP-Mannose Pyrophosphorylase (GDP-MP) is a prominent therapeutic target involved in host-parasite recognition which has been described to be essential for parasite survival. In this work, we produced and purified GDP-MPs from L. mexicana (LmGDP-MP), L. donovani (LdGDP-MP), and human (hGDP-MP), and compared their enzymatic properties. From a rationale design of 100 potential inhibitors, four compounds were identified having a promising and specific inhibitory effect on parasite GDP-MP and antileishmanial activities, one of them exhibits a competitive inhibition on LdGDP-MP and belongs to the 2-substituted quinoline series.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Alkyl-Resorcinol Derivatives as Inhibitors of GDP-Mannose Pyrophosphorylase with Antileishmanial Activities.Molecules. 2021 Mar 11;26(6):1551. doi: 10.3390/molecules26061551. Molecules. 2021. PMID: 33799883 Free PMC article.
-
GDP-Mannose Pyrophosphorylase: A Biologically Validated Target for Drug Development Against Leishmaniasis.Front Cell Infect Microbiol. 2019 May 31;9:186. doi: 10.3389/fcimb.2019.00186. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31214516 Free PMC article. Review.
-
In silico analysis of a therapeutic target in Leishmania infantum: the guanosine-diphospho-D-mannose pyrophosphorylase.Parasite. 2012 Feb;19(1):63-70. doi: 10.1051/parasite/2012191063. Parasite. 2012. PMID: 22314241 Free PMC article.
-
Inhibitors of Leishmania GDP-mannose pyrophosphorylase identified by high-throughput screening of small-molecule chemical library.Antimicrob Agents Chemother. 2010 May;54(5):1712-9. doi: 10.1128/AAC.01634-09. Epub 2010 Feb 16. Antimicrob Agents Chemother. 2010. PMID: 20160053 Free PMC article.
-
Structural insights into the enzymes of the trypanothione pathway: targets for antileishmaniasis drugs.Future Med Chem. 2013 Oct;5(15):1861-75. doi: 10.4155/fmc.13.146. Future Med Chem. 2013. PMID: 24144416 Review.
Cited by
-
Alkyl-Resorcinol Derivatives as Inhibitors of GDP-Mannose Pyrophosphorylase with Antileishmanial Activities.Molecules. 2021 Mar 11;26(6):1551. doi: 10.3390/molecules26061551. Molecules. 2021. PMID: 33799883 Free PMC article.
-
Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.Nat Struct Mol Biol. 2021 May;28(5):1-12. doi: 10.1038/s41594-021-00591-9. Epub 2021 May 13. Nat Struct Mol Biol. 2021. PMID: 33986552
-
UDP-Sugar Producing Pyrophosphorylases: Distinct and Essential Enzymes With Overlapping Substrate Specificities, Providing de novo Precursors for Glycosylation Reactions.Front Plant Sci. 2019 Jan 4;9:1822. doi: 10.3389/fpls.2018.01822. eCollection 2018. Front Plant Sci. 2019. PMID: 30662444 Free PMC article. Review.
-
Minor Impact of A258D Mutation on Biochemical and Enzymatic Properties of Leishmania infantum GDP-Mannose Pyrophosphorylase.Microorganisms. 2022 Jan 21;10(2):231. doi: 10.3390/microorganisms10020231. Microorganisms. 2022. PMID: 35208687 Free PMC article.
-
Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose.Open Life Sci. 2021 Feb 5;16(1):102-107. doi: 10.1515/biol-2021-0015. eCollection 2021. Open Life Sci. 2021. PMID: 33817303 Free PMC article.
References
-
- World Health Organization. Investing to overcome the global impact of neglected tropical diseases. Third WHO report on neglected tropical diseases. WHO/HTM/NTD/2015.1 (2015).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

