Characterization of the Fecal Bacterial Microbiota of Healthy and Diarrheic Dairy Calves
- PMID: 28390070
- PMCID: PMC5435056
- DOI: 10.1111/jvim.14695
Characterization of the Fecal Bacterial Microbiota of Healthy and Diarrheic Dairy Calves
Abstract
Background: Neonatal diarrhea accounts for more than 50% of total deaths in dairy calves. Few population-based studies of cattle have investigated how the microbiota is impacted during diarrhea.
Objectives: To characterize the fecal microbiota and predict the functional potential of the microbial communities in healthy and diarrheic calves.
Methods: Fifteen diarrheic calves between the ages of 1 and 30 days and 15 age-matched healthy control calves were enrolled from 2 dairy farms. The Illumina MiSeq sequencer was used for high-throughput sequencing of the V4 region of the 16S rRNA gene (Illumina, San Diego, CA).
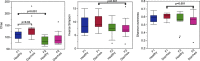
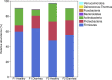
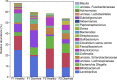
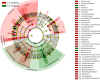
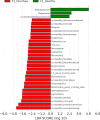
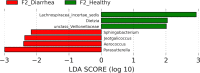
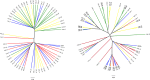
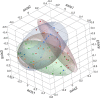
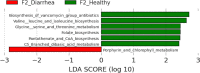
Results: Significant differences in community membership and structure were identified among healthy calves from different farms. Differences in community membership and structure also were identified between healthy and diarrheic calves within each farm. Based on linear discriminant analysis effect size (LEfSe), the genera Bifidobacterium, Megamonas, and a genus of the family Bifidobacteriaceae were associated with health at farm 1, whereas Lachnospiraceae incertae sedis, Dietzia and an unclassified genus of the family Veillonellaceae were significantly associated with health at farm 2. The Phylogenetic Investigation of Communities Reconstruction of Unobserved States (PICRUSt) analysis indicated that diarrheic calves had decreased abundances of genes responsible for metabolism of various vitamins, amino acids, and carbohydrate.
Clinical relevance: The fecal microbiota of healthy dairy calves appeared to be farm specific as were the changes observed during diarrhea. The differences in microbiota structure and membership between healthy and diarrheic calves suggest that dysbiosis can occur in diarrheic calves and it is associated with changes in predictive metagenomic function.
Keywords: Escherichia coli; Bacterial species; Bifidobacterium; Epidemiology; Infectious diseases; LEfSe; Microbiology; PICRUSt.
Copyright © 2017 The Authors. Journal of Veterinary Internal Medicine published by Wiley Periodicals, Inc. on behalf of the American College of Veterinary Internal Medicine.
Figures

Similar articles
-
Dysbiosis of the fecal microbiota in feedlot cattle with hemorrhagic diarrhea.Microb Pathog. 2018 Feb;115:123-130. doi: 10.1016/j.micpath.2017.12.059. Epub 2017 Dec 21. Microb Pathog. 2018. PMID: 29275129
-
Saccharomyces cerevisiae boulardii CNCM I-1079 affects health, growth, and fecal microbiota in milk-fed veal calves.J Dairy Sci. 2019 Aug;102(8):7011-7025. doi: 10.3168/jds.2018-16149. Epub 2019 May 31. J Dairy Sci. 2019. PMID: 31155261
-
Short communication: Comparison of the fecal bacterial communities in diarrheic and nondiarrheic dairy calves from multiple farms in southeastern Pennsylvania.J Dairy Sci. 2021 Jun;104(6):7225-7232. doi: 10.3168/jds.2020-19108. Epub 2021 Mar 2. J Dairy Sci. 2021. PMID: 33663859
-
Diagnostics of dairy and beef cattle diarrhea.Vet Clin North Am Food Anim Pract. 2012 Nov;28(3):443-64. doi: 10.1016/j.cvfa.2012.07.002. Epub 2012 Sep 6. Vet Clin North Am Food Anim Pract. 2012. PMID: 23101670 Free PMC article. Review.
-
Neonatal Calf Diarrhea and Gastrointestinal Microbiota: Etiologic Agents and Microbiota Manipulation for Treatment and Prevention of Diarrhea.Vet Sci. 2024 Feb 29;11(3):108. doi: 10.3390/vetsci11030108. Vet Sci. 2024. PMID: 38535842 Free PMC article. Review.
Cited by
-
Dynamic Changes in Fecal Microbial Communities of Neonatal Dairy Calves by Aging and Diarrhea.Animals (Basel). 2021 Apr 13;11(4):1113. doi: 10.3390/ani11041113. Animals (Basel). 2021. PMID: 33924525 Free PMC article.
-
Effect of Fermented Corn-Soybean Meal on Serum Immunity, the Expression of Genes Related to Gut Immunity, Gut Microbiota, and Bacterial Metabolites in Grower-Finisher Pigs.Front Microbiol. 2019 Nov 20;10:2620. doi: 10.3389/fmicb.2019.02620. eCollection 2019. Front Microbiol. 2019. PMID: 31824447 Free PMC article.
-
Periodical Changes of Feces Microbiota and Its Relationship with Nutrient Digestibility in Early Lambs.Animals (Basel). 2022 Jul 11;12(14):1770. doi: 10.3390/ani12141770. Animals (Basel). 2022. PMID: 35883317 Free PMC article.
-
Varied microbial community assembly and specialization patterns driven by early life microbiome perturbation and modulation in young ruminants.ISME Commun. 2024 Apr 9;4(1):ycae044. doi: 10.1093/ismeco/ycae044. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38650709 Free PMC article.
-
Effects of a farm-specific fecal microbial transplant (FMT) product on clinical outcomes and fecal microbiome composition in preweaned dairy calves.PLoS One. 2022 Oct 21;17(10):e0276638. doi: 10.1371/journal.pone.0276638. eCollection 2022. PLoS One. 2022. PMID: 36269743 Free PMC article.
References
-
- USDA . Dairy 2007, Part I: Reference of Dairy Cattle Health and Management Practices in the United States. Fort Collins CO: USDA‐APHIS‐VS; 2007.
-
- Bäckhed F, Ley RE, Sonnenburg JL, et al. Host‐bacterial mutualism in the human intestine. Science 2005;307:1915–1920. - PubMed
-
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004;4:478–485. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

