Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities
- PMID: 28391084
- PMCID: PMC5777332
- DOI: 10.1016/j.gde.2017.02.004
Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities
Abstract
Mammalian SWI/SNF (BAF) chromatin remodeling complexes orchestrate a diverse set of chromatin alterations which impact transcriptional output. Recent whole-exome sequencing efforts have revealed that the genes encoding subunits of mSWI/SNF complexes are mutated in over 20% of cancers, spanning a wide range of tissue types. The majority of mutations result in loss of subunit protein expression, implicating mSWI/SNF subunits as tumor suppressors. mSWI/SNF-deficient cancers remain a therapeutic challenge, owing to a lack of potent and selective agents which target complexes or unique pathway dependencies generated by mSWI/SNF subunit perturbations. Here, we review the current landscape of mechanistic insights and emerging therapeutic opportunities for human malignancies driven by mSWI/SNF complex perturbation.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
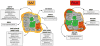

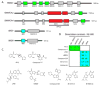
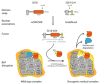
References
-
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. - PubMed
-
- Olins AL, Olins DE. Spheroid chromatin units (ν Bodies) Science. 1974;183:330–332. - PubMed
-
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

